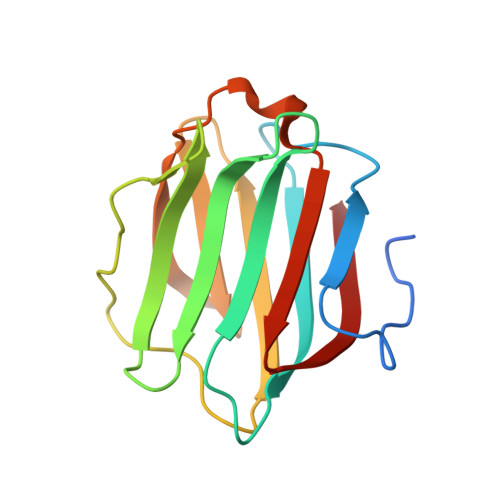
Sterical hindrance promotes selectivity of the autophagy cargo receptor NDP52 for the danger receptor galectin-8 in antibacterial autophagy
Li, S., Wandel, M.P., Li, F., Liu, Z., He, C., Wu, J., Shi, Y., Randow, F.(2013) Sci Signal 6: ra9-ra9
- PubMed: 23386746
- DOI: https://doi.org/10.1126/scisignal.2003730
- Primary Citation of Related Structures:
4GXL - PubMed Abstract:
Autophagy, the process of lysosome-dependent degradation of cytosolic components, is a mechanism by which cells selectively engulf invading pathogens to protect themselves against infection. Galectin-8, a cytosolic protein with specificity for β-galactoside-containing glycans, binds endosomal and lysosomal membranes that have been damaged, for example, by pathogens, and selectively recruits the autophagy cargo receptor NDP52 to induce autophagy. We solved the crystal structure of the NDP52-galectin-8 complex to show how NDP52 exclusively binds galectin-8 and, consequently, why other galectins do not restrict the growth of Salmonella in human cells.
- Hefei National Laboratory for Physical Sciences at Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230026, China.
Organizational Affiliation: