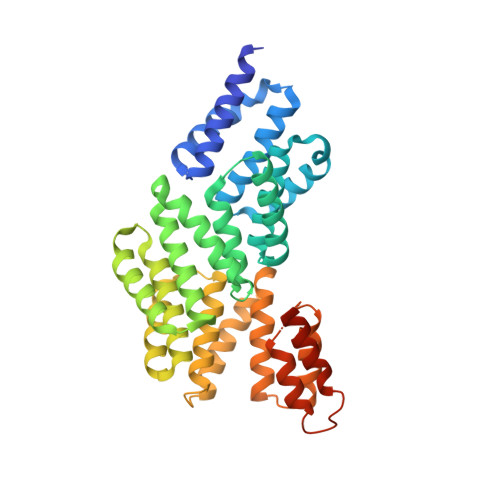
Peptide-binding dependent conformational changes regulate the transcriptional activity of the quorum-sensor NprR.
Zouhir, S., Perchat, S., Nicaise, M., Perez, J., Guimaraes, B., Lereclus, D., Nessler, S.(2013) Nucleic Acids Res 41: 7920-7933
- PubMed: 23793817
- DOI: https://doi.org/10.1093/nar/gkt546
- Primary Citation of Related Structures:
4GPK - PubMed Abstract:
The transcriptional regulator NprR controls the expression of genes essential for the adaptative response of Bacillus cereus. NprR belongs to the RNPP family of directly regulated quorum sensors from Gram-positive bacteria. It is activated by the re-imported signaling peptide NprX. To elucidate the activation mechanism of this quorum-sensing system, we analyzed the conformation changes induced on binding of NprX. We solved the crystal structure of the NprR/NprX binary complex and characterized the apo form of NprR in solution. We demonstrated that apo NprR is a dimer that switches to a tetramer in the presence of NprX. Mutagenesis, and functional analysis allowed us to identify the protein and peptide residues directly involved in the NprR activation process. Based on the comparison with the Rap proteins, we propose a model for the peptide-induced conformational change allowing the apo dimer to switch to an active tetramer specifically recognizing target DNA sequences.
- CNRS, UPR3082, Laboratoire d'Enzymologie et Biochimie Structurales, Gif sur Yvette 91198, France, INRA, UMR1319 Micalis, La Minière, Guyancourt 78280, France, AgroParisTech, UMR1319 Micalis, Jouy-en-Josas 78350, France, Université Paris-Sud, UMR8619, Institut de Biochimie et Biophysique Moléculaire et Cellulaire, Orsay 91405, France and Synchrotron SOLEIL, 91192 Gif sur Yvette, France.
Organizational Affiliation: