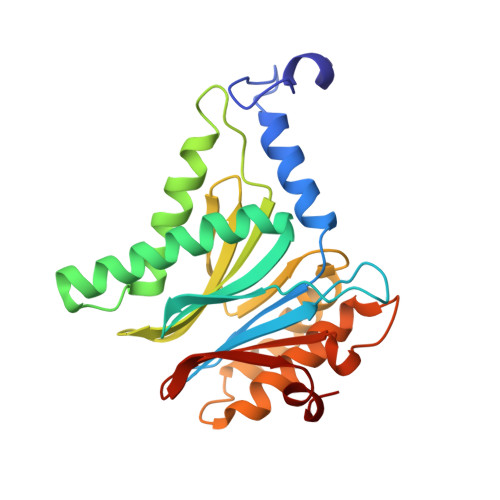
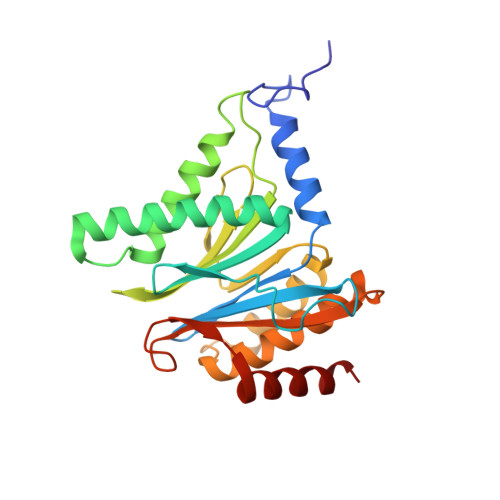
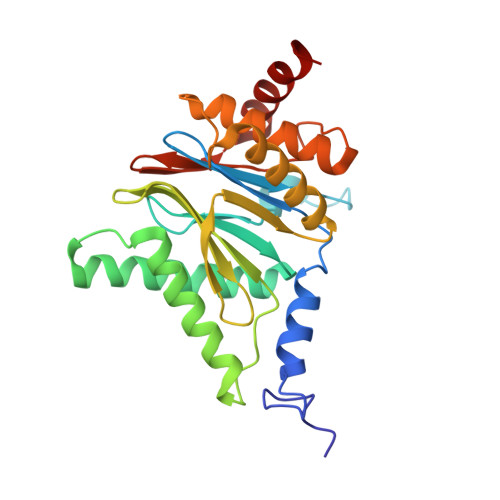
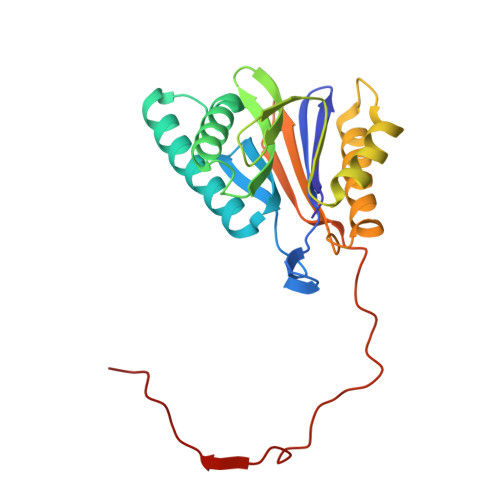
Activity enhancement of the synthetic syrbactin proteasome inhibitor hybrid and biological evaluation in tumor cells.
Archer, C.R., Groll, M., Stein, M.L., Schellenberg, B., Clerc, J., Kaiser, M., Kondratyuk, T.P., Pezzuto, J.M., Dudler, R., Bachmann, A.S.(2012) Biochemistry 51: 6880-6888
- PubMed: 22870914
- DOI: https://doi.org/10.1021/bi300841r
- Primary Citation of Related Structures:
4GK7 - PubMed Abstract:
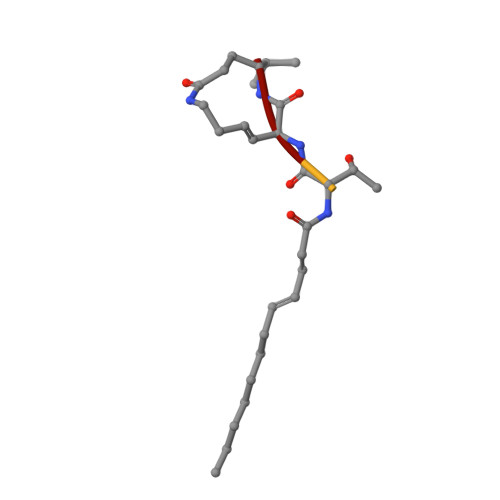
Syrbactins belong to a recently emergent class of bacterial natural product inhibitors that irreversibly inhibit the proteasome of eukaryotes by a novel mechanism. The total syntheses of the syrbactin molecules syringolin A, syringolin B, and glidobactin A have been achieved, which allowed the preparation of syrbactin-inspired derivatives, such as the syringolin A-glidobactin A hybrid molecule (SylA-GlbA). To determine the potency of SylA-GlbA, we employed both in vitro and cell culture-based proteasome assays that measure the subcatalytic chymotrypsin-like (CT-L), trypsin-like (T-L), and caspase-like (C-L) activities. We further studied the inhibitory effects of SylA-GlbA on tumor cell growth using a panel of multiple myeloma, neuroblastoma, and ovarian cancer cell lines and showed that SylA-GlbA strongly blocks the activity of NF-κB. To gain more insights into the structure-activity relationship, we cocrystallized SylA-GlbA in complex with the proteasome and determined the X-ray structure. The electron density map displays covalent binding of the Thr1O(γ) atoms of all active sites to the macrolactam ring of the ligand via ether bond formation, thus providing insights into the structure-activity relationship for the improved affinity of SylA-GlbA for the CT-L activity compared to those of the natural compounds SylA and GlbA. Our study revealed that the novel synthetic syrbactin compound represents one of the most potent proteasome inhibitors analyzed to date and therefore exhibits promising properties for improved drug development as an anticancer therapeutic.
- University of Hawaii Cancer Center, 1236 Lauhala Street, University of Hawaii at Manoa, Honolulu, HI 96813, USA.
Organizational Affiliation: