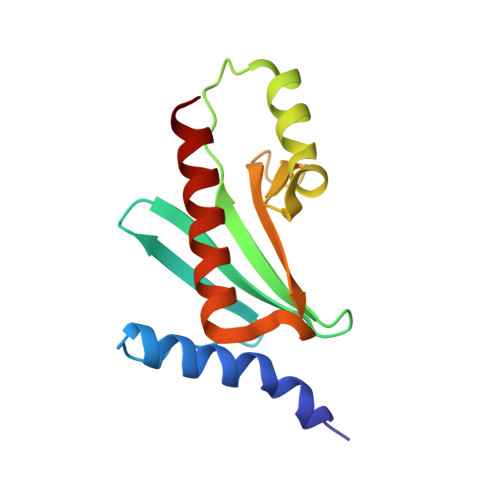
Structure of the HopA1(21-102)-ShcA chaperone-effector complex of Pseudomonas syringae reveals conservation of a virulence factor binding motif from animal to plant pathogens.
Janjusevic, R., Quezada, C.M., Small, J., Stebbins, C.E.(2013) J Bacteriol 195: 658-664
- PubMed: 23204470
- DOI: https://doi.org/10.1128/JB.01621-12
- Primary Citation of Related Structures:
4G6T - PubMed Abstract:
Pseudomonas syringae injects numerous bacterial proteins into host plant cells through a type 3 secretion system (T3SS). One of the first such bacterial effectors discovered, HopA1, is a protein that has unknown functions in the host cell but possesses close homologs that trigger the plant hypersensitive response in resistant strains. Like the virulence factors in many bacterial pathogens of animals, HopA1 depends upon a cognate chaperone in order to be effectively translocated by the P. syringae T3SS. Herein, we report the crystal structure of a complex of HopA1(21-102) with its chaperone, ShcA, determined to 1.56-Å resolution. The structure reveals that three key features of the chaperone-effector interactions found in animal pathogens are preserved in the Gram-negative pathogens of plants, namely, (i) the interaction of the chaperone with a nonglobular polypeptide of the effector, (ii) an interaction centered on the so-called β-motif, and (iii) the presence of a conserved hydrophobic patch in the chaperone that recognizes the β-motif. Structure-based mutagenesis and biochemical studies have established that the β-motif is critical for the stability of this complex. Overall, these results show that the β-motif interactions are broadly conserved in bacterial pathogens utilizing T3SSs, spanning an interkingdom host range.
- Laboratory of Structural Microbiology, The Rockefeller University, New York, NY, USA.
Organizational Affiliation: