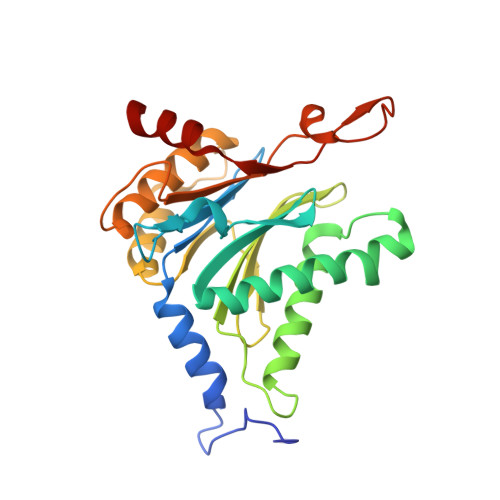
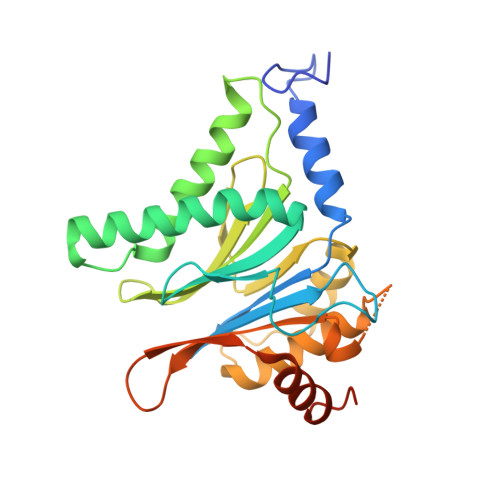
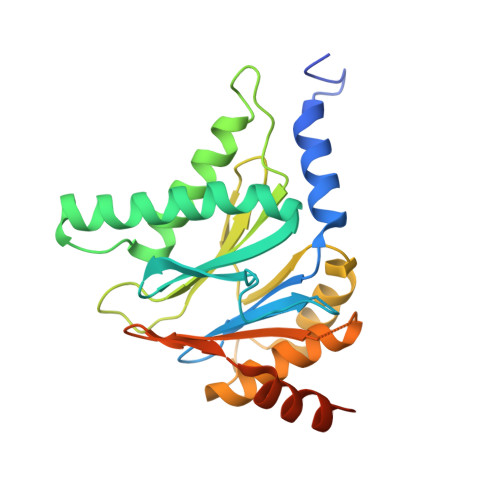
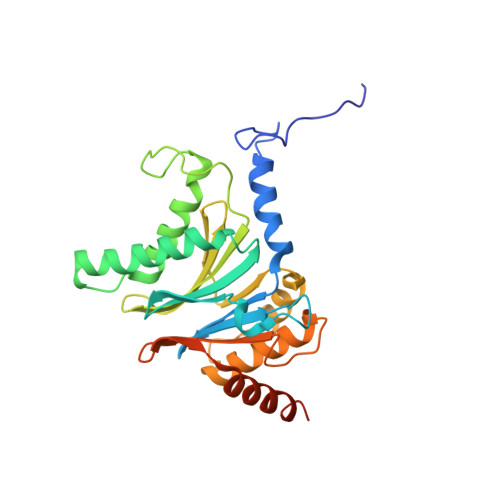
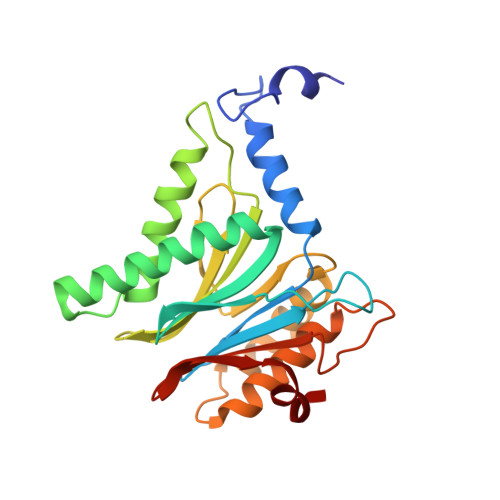
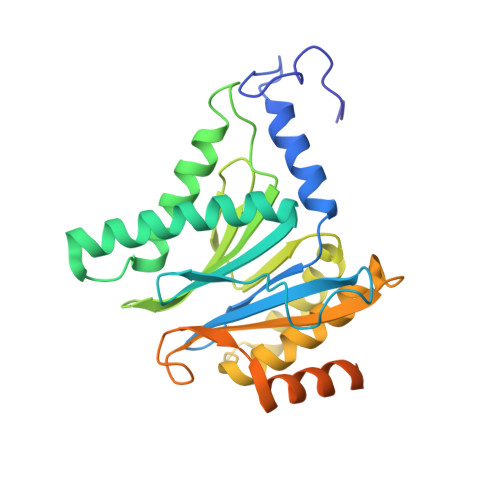
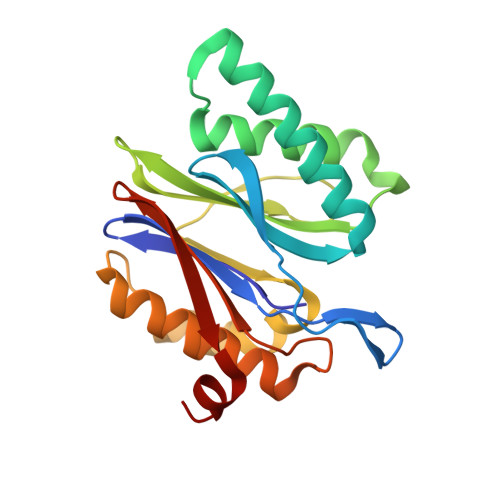
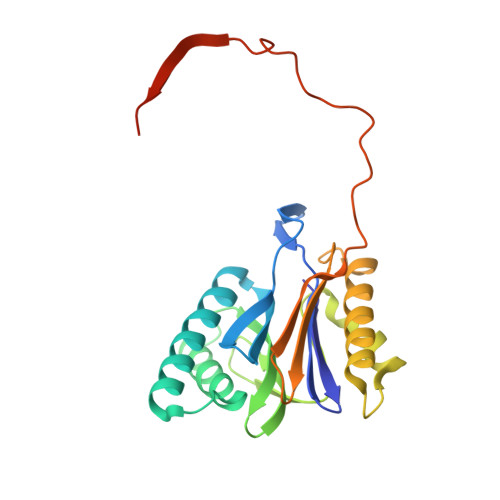
Structure of a Proteasome Pba1-Pba2 Complex: IMPLICATIONS FOR PROTEASOME ASSEMBLY, ACTIVATION, AND BIOLOGICAL FUNCTION.
Stadtmueller, B.M., Kish-Trier, E., Ferrell, K., Petersen, C.N., Robinson, H., Myszka, D.G., Eckert, D.M., Formosa, T., Hill, C.P.(2012) J Biological Chem 287: 37371-37382
- PubMed: 22930756
- DOI: https://doi.org/10.1074/jbc.M112.367003
- Primary Citation of Related Structures:
4G4S - PubMed Abstract:
The 20S proteasome is an essential, 28-subunit protease that sequesters proteolytic sites within a central chamber, thereby repressing substrate degradation until proteasome activators open the entrance/exit gate. Two established activators, Blm10 and PAN/19S, induce gate opening by binding to the pockets between proteasome α-subunits using C-terminal HbYX (hydrophobic-tyrosine-any residue) motifs. Equivalent HbYX motifs have been identified in Pba1 and Pba2, which function in proteasome assembly. Here, we demonstrate that Pba1-Pba2 proteins form a stable heterodimer that utilizes its HbYX motifs to bind mature 20S proteasomes in vitro and that the Pba1-Pba2 HbYX motifs are important for a physiological function of proteasomes, the maintenance of mitochondrial function. Other factors that contribute to proteasome assembly or function also act in the maintenance of mitochondrial function and display complex genetic interactions with one another, possibly revealing an unexpected pathway of mitochondrial regulation involving the Pba1-Pba2 proteasome interaction. Our determination of a proteasome Pba1-Pba2 crystal structure reveals a Pba1 HbYX interaction that is superimposable with those of known activators, a Pba2 HbYX interaction that is different from those reported previously, and a gate structure that is disrupted but not sufficiently open to allow entry of even small peptides. These findings extend understanding of proteasome interactions with HbYX motifs and suggest multiple roles for Pba1-Pba2 interactions throughout proteasome assembly and function.
- Department of Biochemistry, University of Utah School of Medicine, Salt Lake City, Utah 84112-5650, USA.
Organizational Affiliation: