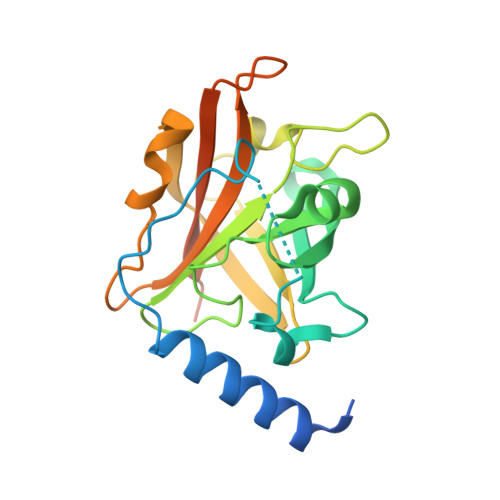
Structural basis for group B streptococcus pilus 1 sortases C regulation and specificity.
Cozzi, R., Prigozhin, D., Rosini, R., Abate, F., Bottomley, M.J., Grandi, G., Telford, J.L., Rinaudo, C.D., Maione, D., Alber, T.(2012) PLoS One 7: e49048-e49048
- PubMed: 23145064
- DOI: https://doi.org/10.1371/journal.pone.0049048
- Primary Citation of Related Structures:
4G1H, 4G1J - PubMed Abstract:
Gram-positive bacteria assemble pili through class C sortase enzymes specialized in polymerizing pilin subunits into covalently linked, high-molecular-weight, elongated structures. Here we report the crystal structures of two class C sortases (SrtC1 and SrtC2) from Group B Streptococcus (GBS) Pilus Island 1. The structures show that both sortases are comprised of two domains: an 8-stranded β-barrel catalytic core conserved among all sortase family members and a flexible N-terminal region made of two α-helices followed by a loop, known as the lid, which acts as a pseudo-substrate. In vitro experiments performed with recombinant SrtC enzymes lacking the N-terminal portion demonstrate that this region of the enzyme is dispensable for catalysis but may have key roles in substrate specificity and regulation. Moreover, in vitro FRET-based assays show that the LPXTG motif common to many sortase substrates is not the sole determinant of sortase C specificity during pilin protein recognition.
- Novartis Vaccines and Diagnostics, Siena, Italy.
Organizational Affiliation: