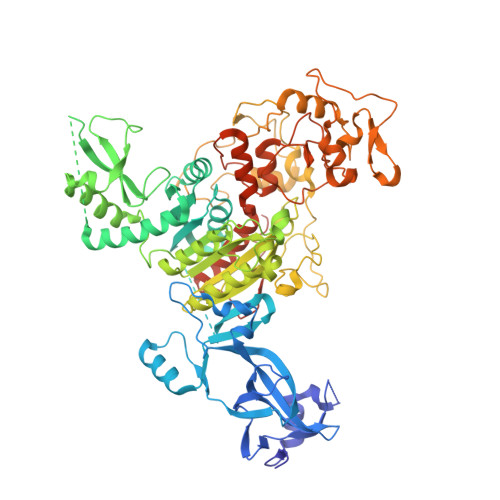
Dual Binding of Chromomethylase Domains to H3K9me2-Containing Nucleosomes Directs DNA Methylation in Plants.
Du, J., Zhong, X., Bernatavichute, Y.V., Stroud, H., Feng, S., Caro, E., Vashisht, A.A., Terragni, J., Chin, H.G., Tu, A., Hetzel, J., Wohlschlegel, J.A., Pradhan, S., Patel, D.J., Jacobsen, S.E.(2012) Cell 151: 167-180
- PubMed: 23021223
- DOI: https://doi.org/10.1016/j.cell.2012.07.034
- Primary Citation of Related Structures:
4FSX, 4FT2, 4FT4 - PubMed Abstract:
DNA methylation and histone modification exert epigenetic control over gene expression. CHG methylation by CHROMOMETHYLASE3 (CMT3) depends on histone H3K9 dimethylation (H3K9me2), but the mechanism underlying this relationship is poorly understood. Here, we report multiple lines of evidence that CMT3 interacts with H3K9me2-containing nucleosomes. CMT3 genome locations nearly perfectly correlated with H3K9me2, and CMT3 stably associated with H3K9me2-containing nucleosomes. Crystal structures of maize CMT3 homolog ZMET2, in complex with H3K9me2 peptides, showed that ZMET2 binds H3K9me2 via both bromo adjacent homology (BAH) and chromo domains. The structures reveal an aromatic cage within both BAH and chromo domains as interaction interfaces that capture H3K9me2. Mutations that abolish either interaction disrupt CMT3 binding to nucleosomes and show a complete loss of CMT3 activity in vivo. Our study establishes dual recognition of H3K9me2 marks by BAH and chromo domains and reveals a distinct mechanism of interplay between DNA methylation and histone modification.
- Structural Biology Program, Memorial Sloan-Kettering Cancer Center, New York, NY 10065, USA.
Organizational Affiliation: