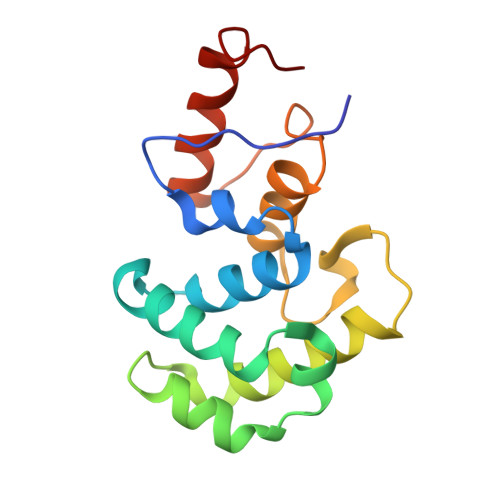
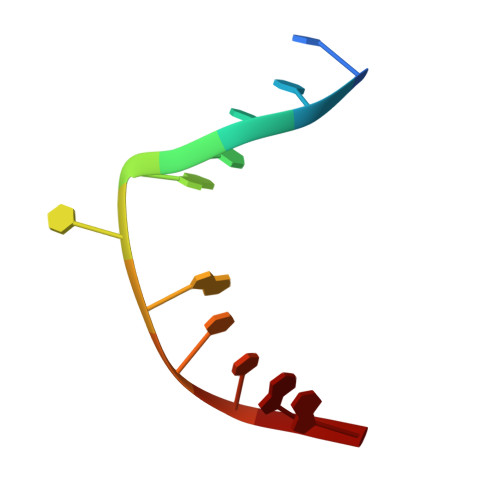
Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Hashimoto, H., Zhang, X., Cheng, X.(2012) Nucleic Acids Res 40: 8276-8284
- PubMed: 22740654
- DOI: https://doi.org/10.1093/nar/gks628
- Primary Citation of Related Structures:
4EVV, 4EW0, 4EW4 - PubMed Abstract:
The mammalian DNA glycosylase--methyl-CpG binding domain protein 4 (MBD4)--is involved in active DNA demethylation via the base excision repair pathway. MBD4 contains an N-terminal MBD and a C-terminal DNA glycosylase domain. MBD4 can excise the mismatched base paired with a guanine (G:X), where X is uracil, thymine or 5-hydroxymethyluracil (5hmU). These are, respectively, the deamination products of cytosine, 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC). Here, we present three structures of the MBD4 C-terminal glycosylase domain (wild-type and its catalytic mutant D534N), in complex with DNA containing a G:T or G:5hmU mismatch. MBD4 flips the target nucleotide from the double-stranded DNA. The catalytic mutant D534N captures the intact target nucleotide in the active site binding pocket. MBD4 specifically recognizes the Watson-Crick polar edge of thymine or 5hmU via the O2, N3 and O4 atoms, thus restricting its activity to thymine/uracil-based modifications while excluding cytosine and its derivatives. The wild-type enzyme cleaves the N-glycosidic bond, leaving the ribose ring in the flipped state, while the cleaved base is released. Unexpectedly, the C1' of the sugar has yet to be hydrolyzed and appears to form a stable intermediate with one of the side chain carboxyl oxygen atoms of D534, via either electrostatic or covalent interaction, suggesting a different catalytic mechanism from those of other DNA glycosylases.
- Department of Biochemistry, Emory University School of Medicine, 1510 Clifton Road, Atlanta, GA 30322, USA.
Organizational Affiliation: