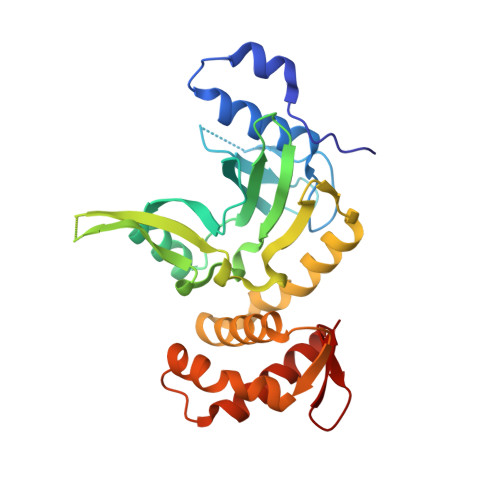
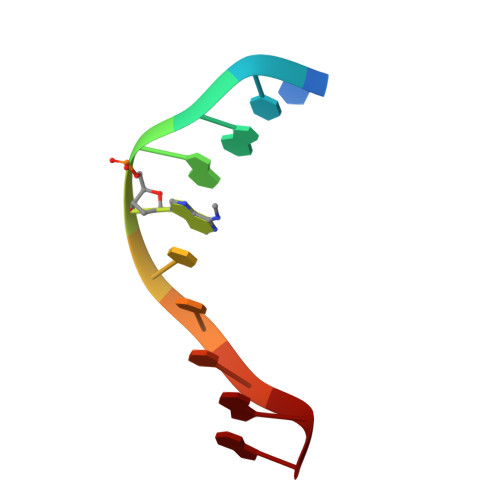
Crystal structure and mechanism of action of the N6-methyladenine-dependent type IIM restriction endonuclease R.DpnI.
Siwek, W., Czapinska, H., Bochtler, M., Bujnicki, J.M., Skowronek, K.(2012) Nucleic Acids Res 40: 7563-7572
- PubMed: 22610857
- DOI: https://doi.org/10.1093/nar/gks428
- Primary Citation of Related Structures:
4ESJ - PubMed Abstract:
DNA methylation-dependent restriction enzymes have many applications in genetic engineering and in the analysis of the epigenetic state of eukaryotic genomes. Nevertheless, high-resolution structures have not yet been reported, and therefore mechanisms of DNA methylation-dependent cleavage are not understood. Here, we present a biochemical analysis and high-resolution DNA co-crystal structure of the N(6)-methyladenine (m6A)-dependent restriction enzyme R.DpnI. Our data show that R.DpnI consists of an N-terminal catalytic PD-(D/E)XK domain and a C-terminal winged helix (wH) domain. Surprisingly, both domains bind DNA in a sequence- and methylation-sensitive manner. The crystal contains R.DpnI with fully methylated target DNA bound to the wH domain, but distant from the catalytic domain. Independent readout of DNA sequence and methylation by the two domains might contribute to R.DpnI specificity or could help the monomeric enzyme to cut the second strand after introducing a nick.
- Laboratory of Bioinformatics and Protein Engineering, International Institute of Molecular and Cell Biology, Trojdena 4, 02-109 Warsaw, Poland.
Organizational Affiliation: