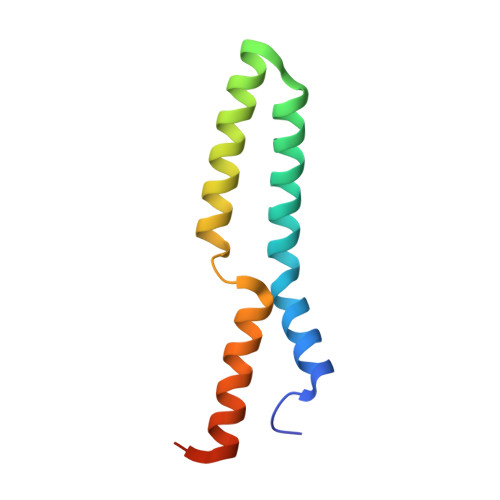
1.55 Angstrom Crystal Structure of the Four Helical Bundle Membrane Localization Domain (4HBM) of the Vibrio vulnificus MARTX Effector Domain DUF5.
Minasov, G., Wawrzak, Z., Geissler, B., Shuvalova, L., Dubrovska, I., Winsor, J., Satchell, K.J., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.