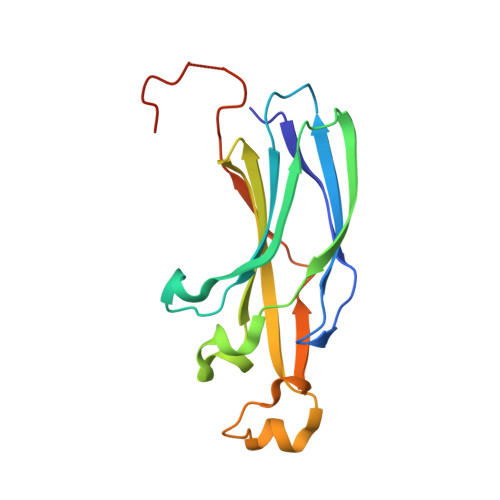
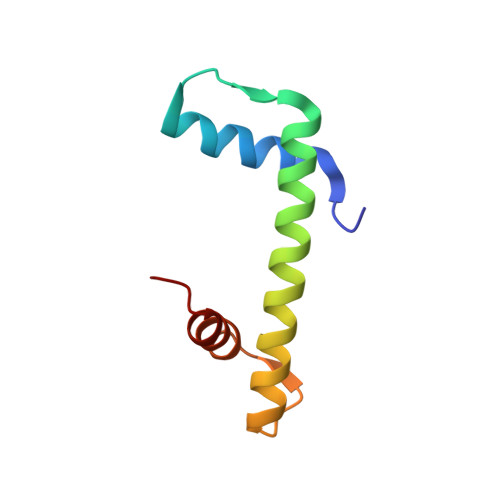
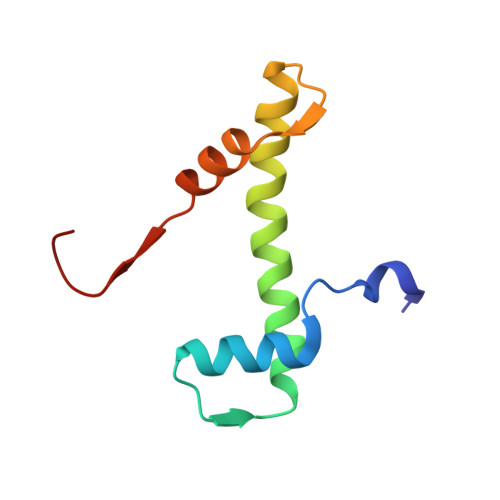
The conformational flexibility of the C-terminus of histone H4 promotes histone octamer and nucleosome stability and yeast viability.
Chavez, M.S., Scorgie, J.K., Dennehey, B.K., Noone, S.M., Tyler, J.K., Churchill, M.E.(2012) Epigenetics Chromatin 5: 5-5
- PubMed: 22541333
- DOI: https://doi.org/10.1186/1756-8935-5-5
- Primary Citation of Related Structures:
4EO5 - PubMed Abstract:
The protein anti-silencing function 1 (Asf1) chaperones histones H3/H4 for assembly into nucleosomes every cell cycle as well as during DNA transcription and repair. Asf1 interacts directly with H4 through the C-terminal tail of H4, which itself interacts with the docking domain of H2A in the nucleosome. The structure of this region of the H4 C-terminus differs greatly in these two contexts. To investigate the functional consequence of this structural change in histone H4, we restricted the available conformations of the H4 C-terminus and analyzed its effect in vitro and in vivo in Saccharomyces cerevisiae. One such mutation, H4 G94P, had modest effects on the interaction between H4 and Asf1. However, in yeast, flexibility of the C-terminal tail of H4 has essential functions that extend beyond chromatin assembly and disassembly. The H4 G94P mutation resulted in severely sick yeast, although nucleosomes still formed in vivo albeit yielding diffuse micrococcal nuclease ladders. In vitro, H4G4P had modest effects on nucleosome stability, dramatically reduced histone octamer stability, and altered nucleosome sliding ability. The functional consequences of altering the conformational flexibility in the C-terminal tail of H4 are severe. Interestingly, despite the detrimental effects of the histone H4 G94P mutant on viability, nucleosome formation was not markedly affected in vivo. However, histone octamer stability and nucleosome stability as well as nucleosome sliding ability were altered in vitro. These studies highlight an important role for correct interactions of the histone H4 C-terminal tail within the histone octamer and suggest that maintenance of a stable histone octamer in vivo is an essential feature of chromatin dynamics.
- Department of Biochemistry and Molecular Biology, University of Texas MD Anderson Cancer Center, Houston, TX, 77030, USA.
Organizational Affiliation: