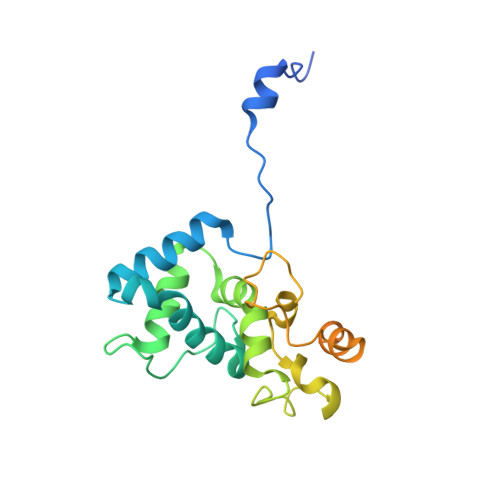
Crystal structure of the coat protein of the flexible filamentous papaya mosaic virus.
Yang, S., Wang, T., Bohon, J., Gagne, N., Bolduc, M., Leclerc, D., Li, H.(2012) J Mol Biology 422: 263-273
- PubMed: 22659319
- DOI: https://doi.org/10.1016/j.jmb.2012.05.032
- Primary Citation of Related Structures:
4DOX - PubMed Abstract:
Papaya mosaic virus (PapMV) is a filamentous plant virus that belongs to the Alphaflexiviridae family. Flexible filamentous viruses have defied more than two decades of effort in fiber diffraction, and no high-resolution structure is available for any member of the Alphaflexiviridae family. Here, we report our structural characterization of PapMV by X-ray crystallography and cryo-electron microscopy three-dimensional reconstruction. We found that PapMV is 135Å in diameter with a helical symmetry of ~10 subunits per turn. Crystal structure of the C-terminal truncated PapMV coat protein (CP) reveals a novel all-helix fold with seven α-helices. Thus, the PapMVCP structure is different from the four-helix-bundle fold of tobacco mosaic virus in which helix bundling dominates the subunit interface in tobacco mosaic virus and conveys rigidity to the rod virus. PapMV CP was crystallized as an asymmetrical dimer in which one protein lassoes the other by the N-terminal peptide. Mutation of residues critical to the inter-subunit lasso interaction abolishes CP polymerization. The crystal structure suggests that PapMV may polymerize via the consecutive N-terminal loop lassoing mechanism. The structure of PapMV will be useful for rational design and engineering of the PapMV nanoparticles into innovative vaccines.
- Biology Department, Brookhaven National Laboratory, Upton, NY 11973, USA.
Organizational Affiliation: