Synthesis and Screening of a Haloacetamidine Containing Library To Identify PAD4 Selective Inhibitors.
Jones, J.E., Slack, J.L., Fang, P., Zhang, X., Subramanian, V., Causey, C.P., Coonrod, S.A., Guo, M., Thompson, P.R.(2012) ACS Chem Biol 7: 160-165
- PubMed: 22004374
- DOI: https://doi.org/10.1021/cb200258q
- Primary Citation of Related Structures:
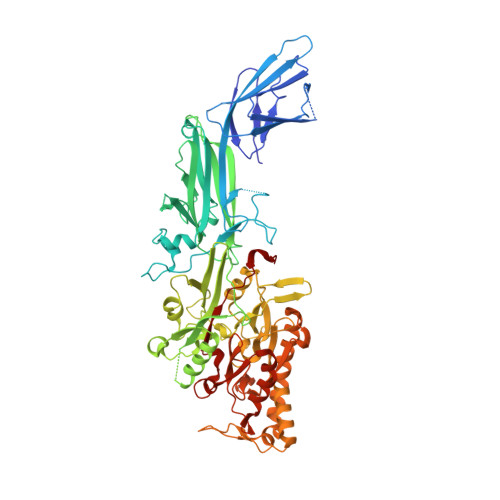
4DKT - PubMed Abstract:
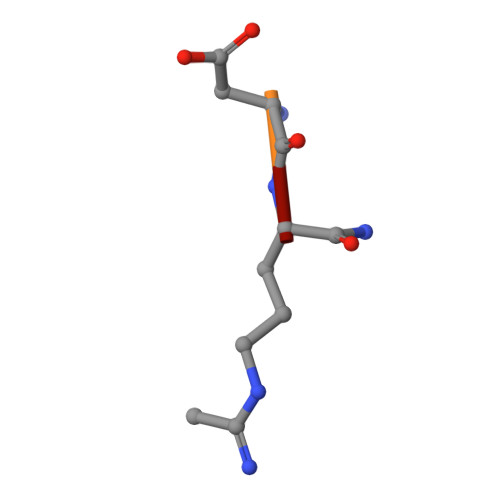
Protein arginine deiminase activity (PAD) is increased in cancer, rheumatoid arthritis, and ulcerative colitis. Although the link between abnormal PAD activity and disease is clear, the relative contribution of the individual PADs to human disease is not known; there are 5 PAD isozymes in humans. Building on our previous development of F- and Cl-amidine as potent pan-PAD irreversible inhibitors, we describe herein a library approach that was used to identify PAD-selective inhibitors. Specifically, we describe the identification of Thr-Asp-F-amidine (TDFA) as a highly potent PAD4 inactivator that displays ≥15-fold selectivity for PAD4 versus PAD1 and ≥50-fold versus PADs 2 and 3. This compound is active in cells and can be used to inhibit PAD4 activity in cellulo. The structure of the PAD4·TDFA complex has also been solved, and the structure and mutagenesis data indicate that the enhanced potency is due to interactions between the side chains of Q346, R374, and R639. Finally, we converted TDFA into a PAD4-selective ABPP and demonstrated that this compound, biotin-TDFA, can be used to selectively isolate purified PAD4 in vitro. In total, TDFA and biotin-TDFA represent PAD4-selective chemical probes that can be used to study the physiological roles of this enzyme.