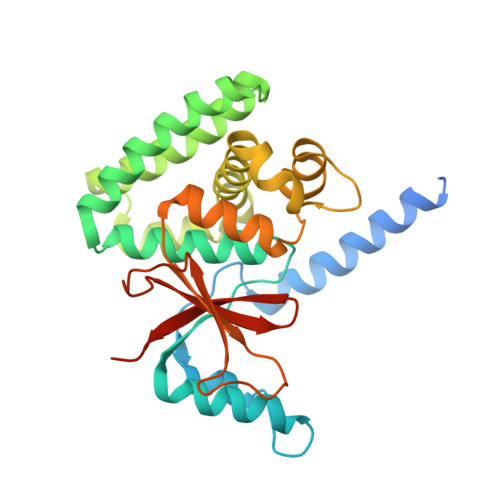
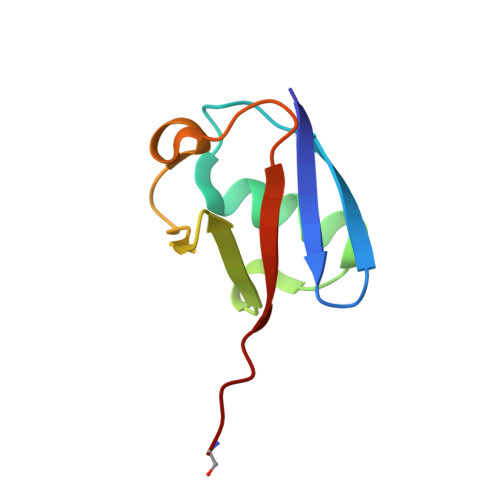
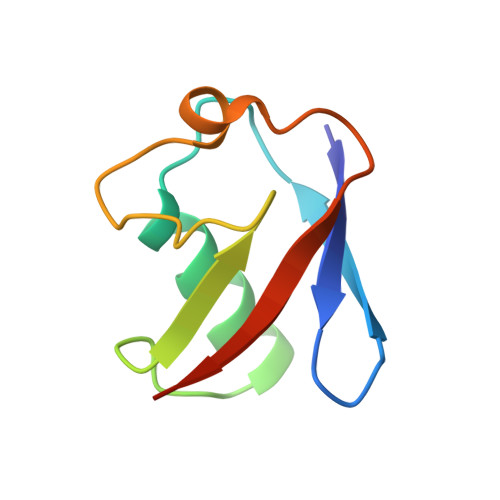
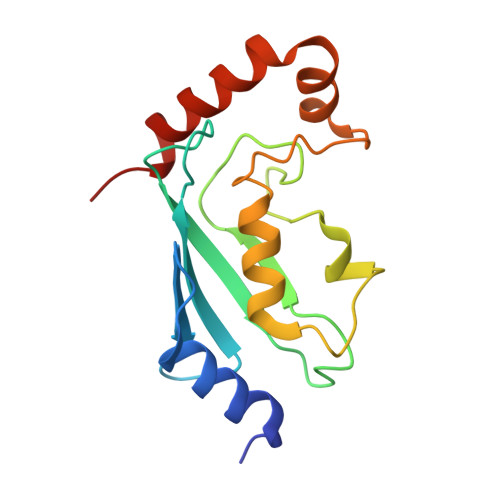
The mechanism of OTUB1-mediated inhibition of ubiquitination.
Wiener, R., Zhang, X., Wang, T., Wolberger, C.(2012) Nature 483: 618-622
- PubMed: 22367539
- DOI: https://doi.org/10.1038/nature10911
- Primary Citation of Related Structures:
4DHI, 4DHJ, 4DHZ - PubMed Abstract:
Histones are ubiquitinated in response to DNA double-strand breaks (DSB), promoting recruitment of repair proteins to chromatin. UBC13 (also known as UBE2N) is a ubiquitin-conjugating enzyme (E2) that heterodimerizes with UEV1A (also known as UBE2V1) and synthesizes K63-linked polyubiquitin (K63Ub) chains at DSB sites in concert with the ubiquitin ligase (E3), RNF168 (ref. 3). K63Ub synthesis is regulated in a non-canonical manner by the deubiquitinating enzyme, OTUB1 (OTU domain-containing ubiquitin aldehyde-binding protein 1), which binds preferentially to the UBC13∼Ub thiolester. Residues amino-terminal to the OTU domain, which had been implicated in ubiquitin binding, are required for binding to UBC13∼Ub and inhibition of K63Ub synthesis. Here we describe structural and biochemical studies elucidating how OTUB1 inhibits UBC13 and other E2 enzymes. We unexpectedly find that OTUB1 binding to UBC13∼Ub is allosterically regulated by free ubiquitin, which binds to a second site in OTUB1 and increases its affinity for UBC13∼Ub, while at the same time disrupting interactions with UEV1A in a manner that depends on the OTUB1 N terminus. Crystal structures of an OTUB1-UBC13 complex and of OTUB1 bound to ubiquitin aldehyde and a chemical UBC13∼Ub conjugate show that binding of free ubiquitin to OTUB1 triggers conformational changes in the OTU domain and formation of a ubiquitin-binding helix in the N terminus, thus promoting binding of the conjugated donor ubiquitin in UBC13∼Ub to OTUB1. The donor ubiquitin thus cannot interact with the E2 enzyme, which has been shown to be important for ubiquitin transfer. The N-terminal helix of OTUB1 is positioned to interfere with UEV1A binding to UBC13, as well as with attack on the thiolester by an acceptor ubiquitin, thereby inhibiting K63Ub synthesis. OTUB1 binding also occludes the RING E3 binding site on UBC13, thus providing a further component of inhibition. The general features of the inhibition mechanism explain how OTUB1 inhibits other E2 enzymes in a non-catalytic manner.
- Department of Biophysics and Biophysical Chemistry and Howard Hughes Medical Institute, Johns Hopkins University School of Medicine, Baltimore, Maryland 21205, USA.
Organizational Affiliation: