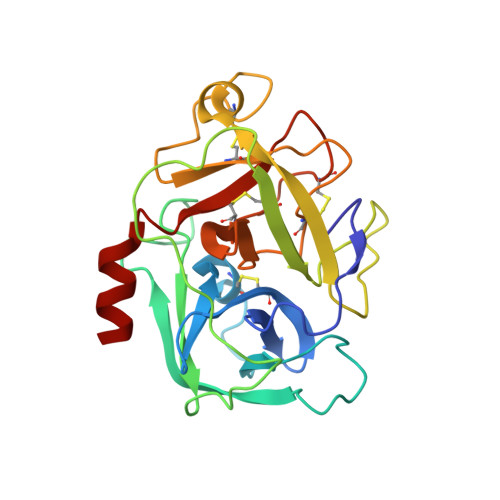
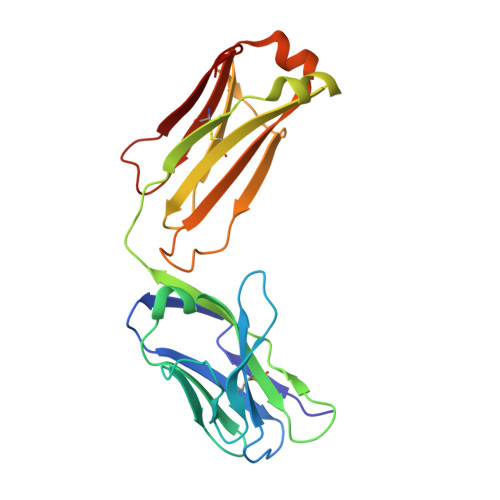
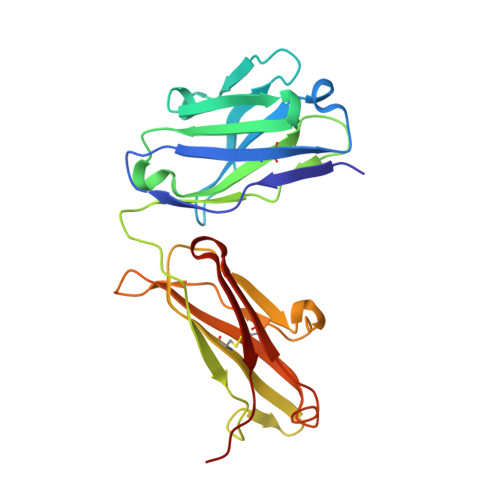
Inhibiting alternative pathway complement activation by targeting the factor d exosite.
Katschke, K.J., Wu, P., Ganesan, R., Kelley, R.F., Mathieu, M.A., Hass, P.E., Murray, J., Kirchhofer, D., Wiesmann, C., van Lookeren Campagne, M.(2012) J Biological Chem 287: 12886-12892
- PubMed: 22362762
- DOI: https://doi.org/10.1074/jbc.M112.345082
- Primary Citation of Related Structures:
4D9Q, 4D9R - PubMed Abstract:
By virtue of its amplifying property, the alternative complement pathway has been implicated in a number of inflammatory diseases and constitutes an attractive therapeutic target. An anti-factor D Fab fragment (AFD) was generated to inhibit the alternative complement pathway in advanced dry age-related macular degeneration. AFD potently prevented factor D (FD)-mediated proteolytic activation of its macromolecular substrate C3bB, but not proteolysis of a small synthetic substrate, indicating that AFD did not block access of the substrate to the catalytic site. The crystal structures of AFD in complex with human and cynomolgus FD (at 2.4 and 2.3 Å, respectively) revealed the molecular details of the inhibitory mechanism. The structures show that the AFD-binding site includes surface loops of FD that form part of the FD exosite. Thus, AFD inhibits FD proteolytic function by interfering with macromolecular substrate access rather than by inhibiting FD catalysis, providing the molecular basis of AFD-mediated inhibition of a rate-limiting step in the alternative complement pathway.
- Department of Immunology, Genentech Incorporated, South San Francisco, California 94080, USA.
Organizational Affiliation: