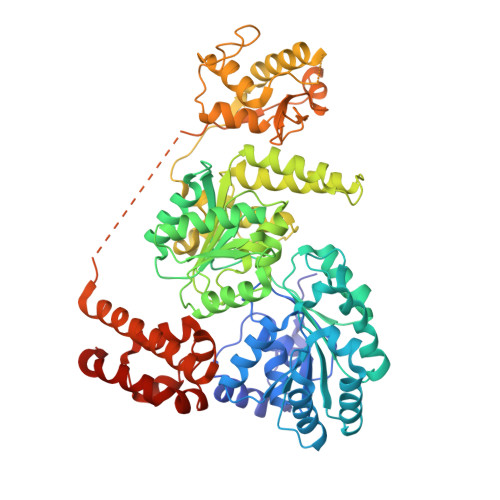
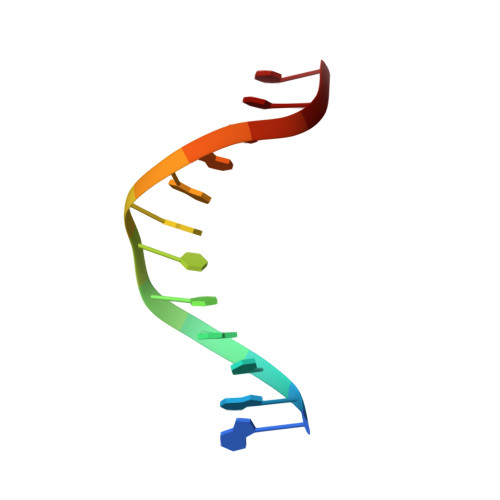
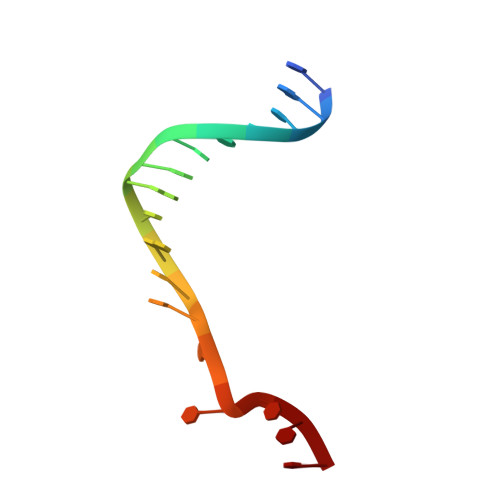
Crystal Structure of the Bloom'S Syndrome Helicase Indicates a Role for the Hrdc Domain in Conformational Changes.
Newman, J.A., Savitsky, P., Allerston, C.K., Bizard, A.H., Ozer, O., Sarlos, K., Liu, Y., Pardon, E., Steyaert, J., Hickson, I.D., Gileadi, O.(2015) Nucleic Acids Res 43: 5221
- PubMed: 25901030
- DOI: https://doi.org/10.1093/nar/gkv373
- Primary Citation of Related Structures:
4CDG, 4CGZ - PubMed Abstract:
Bloom's syndrome helicase (BLM) is a member of the RecQ family of DNA helicases, which play key roles in the maintenance of genome integrity in all organism groups. We describe crystal structures of the BLM helicase domain in complex with DNA and with an antibody fragment, as well as SAXS and domain association studies in solution. We show an unexpected nucleotide-dependent interaction of the core helicase domain with the conserved, poorly characterized HRDC domain. The BLM-DNA complex shows an unusual base-flipping mechanism with unique positioning of the DNA duplex relative to the helicase core domains. Comparison with other crystal structures of RecQ helicases permits the definition of structural transitions underlying ATP-driven helicase action, and the identification of a nucleotide-regulated tunnel that may play a role in interactions with complex DNA substrates.
- Structural Genomics Consortium, University of Oxford, ORCRB, Roosevelt Drive, Oxford OX3 7DQ, UK.
Organizational Affiliation: