Structural Basis of Pcsb-Mediated Cell Separation in Streptococcus Pneumoniae.
Bartual, S.G., Straume, D., Stamsas, G.A., Munoz, I.G., Alfonso, C., Martinez-Ripoll, M., Havarstein, L.S., Hermoso, J.A.(2014) Nat Commun 5: 3842
- PubMed: 24804636
- DOI: https://doi.org/10.1038/ncomms4842
- Primary Citation of Related Structures:
4CGK - PubMed Abstract:
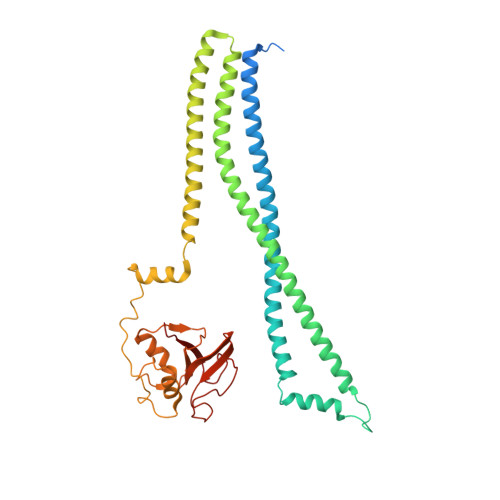
Separation of daughter cells during bacterial cell division requires splitting of the septal cross wall by peptidoglycan hydrolases. In Streptococcus pneumoniae, PcsB is predicted to perform this operation. Recent evidence shows that PcsB is recruited to the septum by the transmembrane FtsEX complex, and that this complex is required for cell division. However, PcsB lacks detectable catalytic activity in vitro, and while it has been proposed that FtsEX activates PcsB, evidence for this is lacking. Here we demonstrate that PcsB has muralytic activity, and report the crystal structure of full-length PcsB. The protein adopts a dimeric structure in which the V-shaped coiled-coil (CC) domain of each monomer acts as a pair of molecular tweezers locking the catalytic domain of each dimeric partner in an inactive configuration. This suggests that the release of the catalytic domains likely requires an ATP-driven conformational change in the FtsEX complex, conveyed towards the catalytic domains through coordinated movements of the CC domain.
- 1] Department of Crystallography and Structural Biology, Instituto de Química-Física Rocasolano, CSIC, Serrano 119, 28006 Madrid, Spain [2].
Organizational Affiliation: