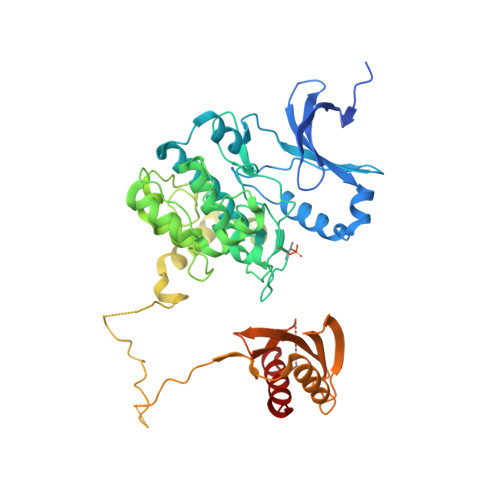
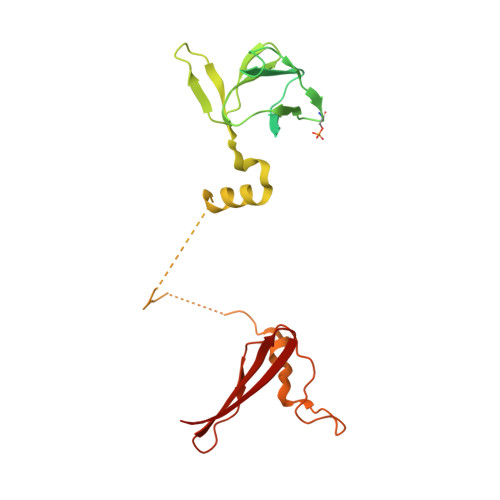
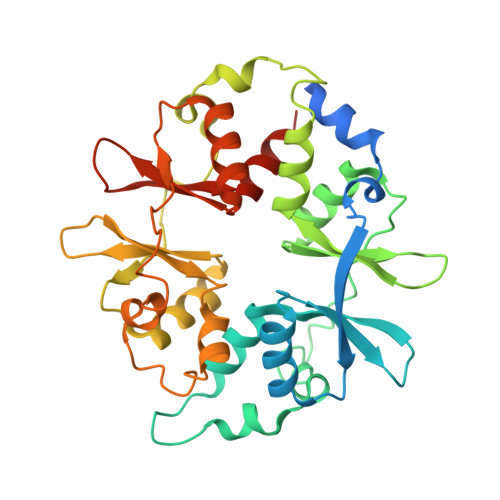
Structural Basis of Ampk Regulation by Small Molecule Activators.
Xiao, B., Sanders, M.J., Carmena, D., Bright, N.J., Haire, L.F., Underwood, E., Patel, B.R., Heath, R.B., Walker, P.A., Hallen, S., Giordanetto, F., Martin, S.R., Carling, D., Gamblin, S.J.(2013) Nat Commun 4: 3017
- PubMed: 24352254
- DOI: https://doi.org/10.1038/ncomms4017
- Primary Citation of Related Structures:
4CFE, 4CFF - PubMed Abstract:
AMP-activated protein kinase (AMPK) plays a major role in regulating cellular energy balance by sensing and responding to increases in AMP/ADP concentration relative to ATP. Binding of AMP causes allosteric activation of the enzyme and binding of either AMP or ADP promotes and maintains the phosphorylation of threonine 172 within the activation loop of the kinase. AMPK has attracted widespread interest as a potential therapeutic target for metabolic diseases including type 2 diabetes and, more recently, cancer. A number of direct AMPK activators have been reported as having beneficial effects in treating metabolic diseases, but there has been no structural basis for activator binding to AMPK. Here we present the crystal structure of human AMPK in complex with a small molecule activator that binds at a site between the kinase domain and the carbohydrate-binding module, stabilising the interaction between these two components. The nature of the activator-binding pocket suggests the involvement of an additional, as yet unidentified, metabolite in the physiological regulation of AMPK. Importantly, the structure offers new opportunities for the design of small molecule activators of AMPK for treatment of metabolic disorders.
- 1] MRC National Institute for Medical Research, The Ridgeway, Mill Hill, London NW7 1AA, UK [2].
Organizational Affiliation: