Architecture of the Lsm1-7-Pat1 Complex: A Conserved Assembly in Eukaryotic Mrna Turnover
Sharif, H., Conti, E.(2013) Cell Rep 5: 283
- PubMed: 24139796
- DOI: https://doi.org/10.1016/j.celrep.2013.10.004
- Primary Citation of Related Structures:
4C8Q, 4C92 - PubMed Abstract:
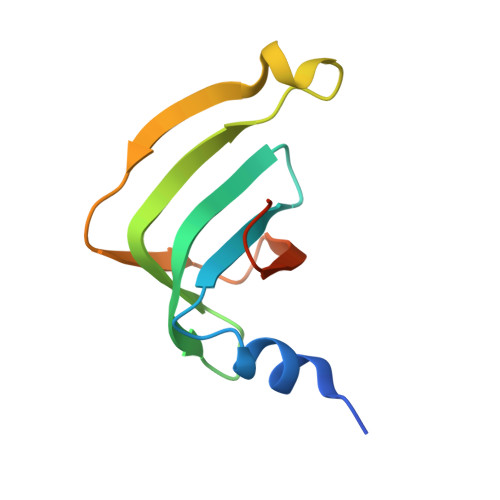
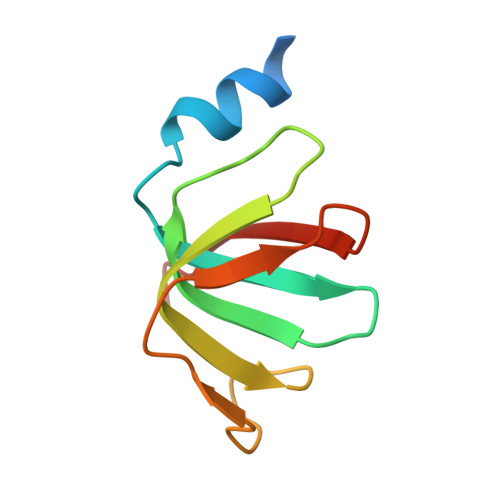
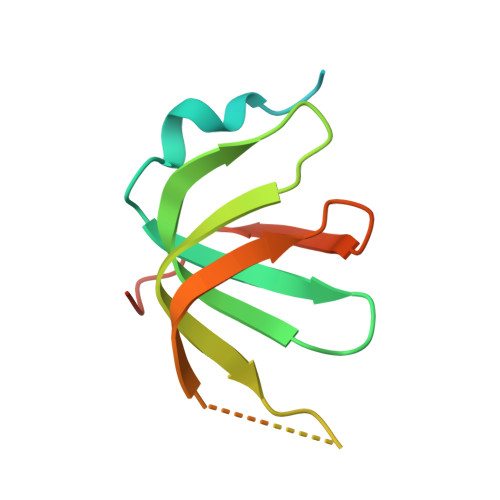
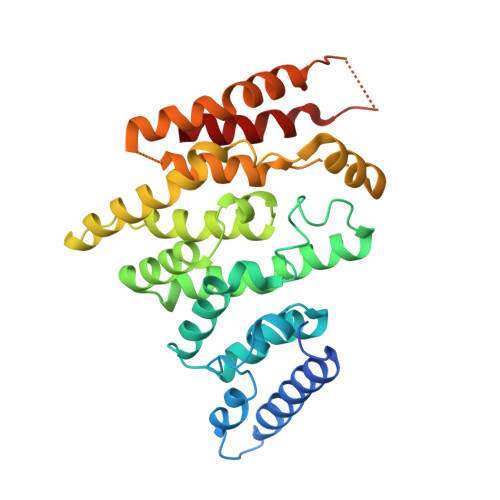
The decay of mRNAs is a key step in eukaryotic gene expression. The cytoplasmic Lsm1-7-Pat1 complex is a conserved component of the 5'-to-3' mRNA decay pathway, linking deadenylation to decapping. Lsm1-7 is similar to the nuclear Sm complexes that bind oligo-uridine tracts in snRNAs. The 2.3 Å resolution structure of S. cerevisiae Lsm1-7 shows the presence of a heptameric ring with Lsm1-2-3-6-5-7-4 topology. A distinct structural feature of the cytoplasmic Lsm ring is the C-terminal extension of Lsm1, which plugs the exit site of the central channel and approaches the RNA binding pockets. The 3.7 Å resolution structure of Lsm1-7 bound to the C-terminal domain of Pat1 reveals that Pat1 recognition is not mediated by the distinguishing cytoplasmic subunit, Lsm1, but by Lsm2 and Lsm3. These results show how the auxiliary domains and the canonical Sm folds of the Lsm1-7 complex are organized in order to mediate and modulate macromolecular interactions.
- Department of Structural Cell Biology, Max Planck Institute of Biochemistry, Am Klopferspitz 18, 82152 Martinsried, Germany.
Organizational Affiliation: