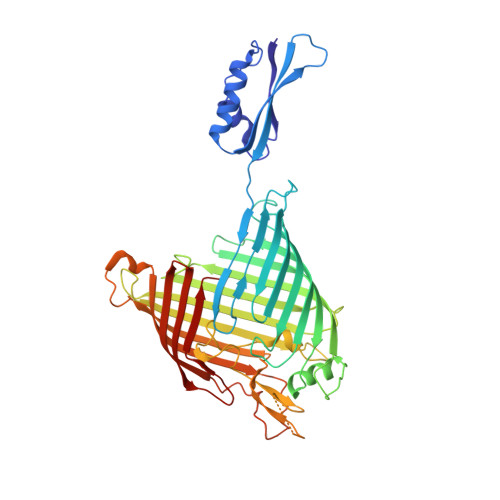
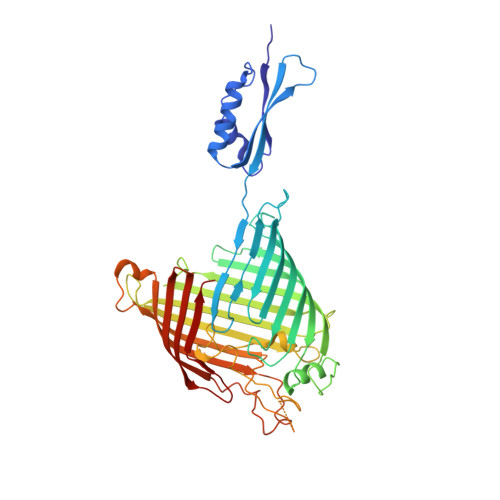
Structure of Bama, an Essential Factor in Outer Membrane Protein Biogenesis
Albrecht, R., Schuetz, M., Oberhettinger, P., Faulstich, M., Bermejo, I., Rudel, T., Diederichs, K., Zeth, K.(2014) Acta Crystallogr D Biol Crystallogr 70: 1779
- PubMed: 24914988
- DOI: https://doi.org/10.1107/S1399004714007482
- Primary Citation of Related Structures:
4C4V - PubMed Abstract:
Outer membrane protein (OMP) biogenesis is an essential process for maintaining the bacterial cell envelope and involves the β-barrel assembly machinery (BAM) for OMP recognition, folding and assembly. In Escherichia coli this function is orchestrated by five proteins: the integral outer membrane protein BamA of the Omp85 superfamily and four associated lipoproteins. To unravel the mechanism underlying OMP folding and insertion, the structure of the E. coli BamA β-barrel and P5 domain was determined at 3 Å resolution. These data add information beyond that provided in the recently published crystal structures of BamA from Haemophilus ducreyi and Neisseria gonorrhoeae and are a valuable basis for the interpretation of pertinent functional studies. In an `open' conformation, E. coli BamA displays a significant degree of flexibility between P5 and the barrel domain, which is indicative of a multi-state function in substrate transfer. E. coli BamA is characterized by a discontinuous β-barrel with impaired β1-β16 strand interactions denoted by only two connecting hydrogen bonds and a disordered C-terminus. The 16-stranded barrel surrounds a large cavity which implies a function in OMP substrate binding and partial folding. These findings strongly support a mechanism of OMP biogenesis in which substrates are partially folded inside the barrel cavity and are subsequently released laterally into the lipid bilayer.
- Department of Protein Evolution, Max-Planck-Institut für Entwicklungsbiologie, Spemannstrasse 35, 72076 Tübingen, Germany.
Organizational Affiliation: