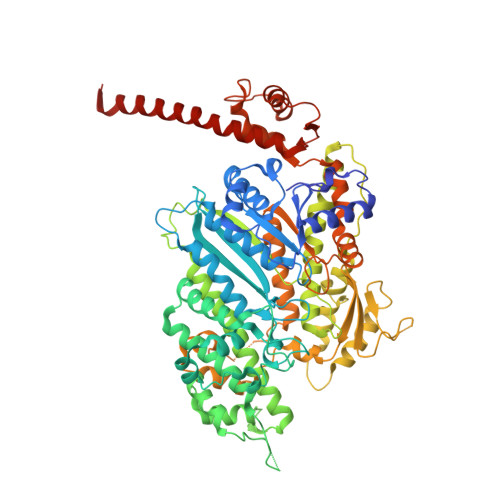
Crystal Structure of Human Myosin 1C-the Motor in Glut4 Exocytosis: Implications for Ca(2+) Regulation and 14-3-3 Binding.
Munnich, S., Taft, M.H., Manstein, D.J.(2014) J Mol Biology 426: 2070
- PubMed: 24636949
- DOI: https://doi.org/10.1016/j.jmb.2014.03.004
- Primary Citation of Related Structures:
4BYF - PubMed Abstract:
Myosin 1c (Myo1c) plays a key role in supporting motile events that underlie cell migration, vesicle trafficking, insulin-stimulated glucose uptake and hearing. Here, we present the crystal structure of the human Myo1c motor in complex with its light chain calmodulin. Our structure reveals tight interactions of the motor domain with calmodulin bound to the first IQ motif in the neck region. Several of the calmodulin residues contributing to this interaction are also involved in Ca(2+) binding. Contact residues in the motor domain are linked to the central β-sheet and the HO helix, suggesting a mechanism for communicating changes in Ca(2+) binding in the neck region to the actin and nucleotide binding regions of the motor domain. The structural context and the chemical environment of Myo1c mutations that are involved in sensorineural hearing loss in humans are described and their impact on motor function is discussed. We show that a construct consisting of the motor domain of Myo1c and the first IQ motif is sufficient to establish a tight interaction with 14-3-3β (KD=0.9 μM) and present the model of a double-headed Myo1c-14-3-3 complex. This complex has been implicated in the exocytosis of glucose transporter 4 storage vesicles during insulin-stimulated glucose uptake.
- Institute for Biophysical Chemistry, Hannover Medical School, Carl-Neuberg-Str. 1, 30625 Hannover, Germany.
Organizational Affiliation: