Small Peptides Blocking Inhibition of Factor Xa and Tissue Factor-Factor Viia by Tissue Factor Pathway Inhibitor (Tfpi)
Dockal, M., Hartman, R., Fries, M., Christella, M., Thomassen, L.G.D., Heizman, A., Ehrlich, H., Rosing, J., Osterkamp, F., Polakowski, T., Reineke, U., Griessner, A., Brandstetter, H., Scheiflinger, F.(2014) J Biological Chem 289: 1732
- PubMed: 24275667
- DOI: https://doi.org/10.1074/jbc.M113.533836
- Primary Citation of Related Structures:
4BQD - PubMed Abstract:
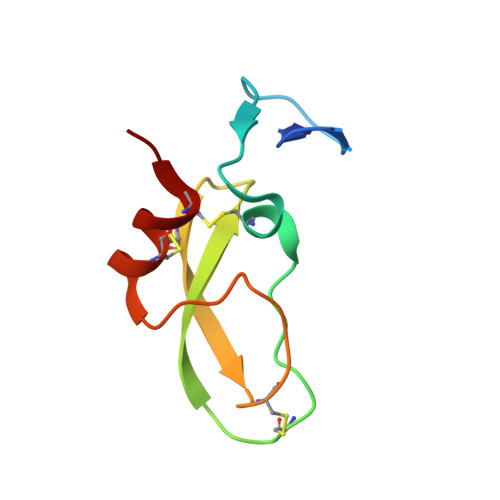
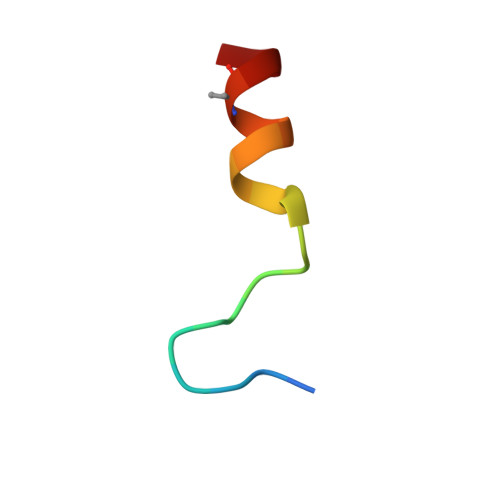
Tissue factor pathway inhibitor (TFPI) is a Kunitz-type protease inhibitor that inhibits activated factor X (FXa) via a slow-tight binding mechanism and tissue factor-activated FVII (TF-FVIIa) via formation of a quaternary FXa-TFPI-TF-FVIIa complex. Inhibition of TFPI enhances coagulation in hemophilia models. Using a library approach, we selected and subsequently optimized peptides that bind TFPI and block its anticoagulant activity. One peptide (termed compound 3), bound with high affinity to the Kunitz-1 (K1) domain of TFPI (Kd ∼1 nM). We solved the crystal structure of this peptide in complex with the K1 of TFPI at 2.55-Å resolution. The structure of compound 3 can be segmented into a N-terminal anchor; an Ω-shaped loop; an intermediate segment; a tight glycine-loop; and a C-terminal α-helix that is anchored to K1 at its reactive center loop and two-stranded β-sheet. The contact surface has an overall hydrophobic character with some charged hot spots. In a model system, compound 3 blocked FXa inhibition by TFPI (EC50 = 11 nM) and inhibition of TF-FVIIa-catalyzed FX activation by TFPI (EC50 = 2 nM). The peptide prevented transition from the loose to the tight FXa-TFPI complex, but did not affect formation of the loose FXa-TFPI complex. The K1 domain of TFPI binds and inhibits FVIIa and the K2 domain similarly inhibits FXa. Because compound 3 binds to K1, our data show that K1 is not only important for FVIIa inhibition but also for FXa inhibition, i.e. for the transition of the loose to the tight FXa-TFPI complex. This mode of action translates into normalization of coagulation of hemophilia plasmas. Compound 3 thus bears potential to prevent bleeding in hemophilia patients.
- From Baxter Innovations GmbH, Uferstrasse 15, A-2304 Orth/Donau, Austria.
Organizational Affiliation: