The Solution Structure and Dynamics of Full-Length Human Cerebral Dopamine Neurotrophic Factor and its Neuroprotective Role Against Alpha-Synuclein Oligomers.
Latge, C., Cabral, K.M.S., De Oliveira, G.A.P., Raymundo, D.P., Freitas, J.A., Johanson, L., Romao, L.F., Palhano, F.L., Herrmann, T., Almeida, M.S., Foguel, D.(2015) J Biological Chem 290: 20527-20540
- PubMed: 26149686
- DOI: https://doi.org/10.1074/jbc.M115.662254
- Primary Citation of Related Structures:
4BIT - PubMed Abstract:
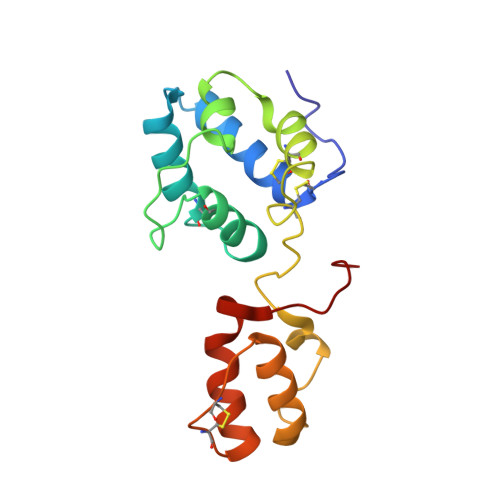
Cerebral dopamine neurotrophic factor (CDNF) is a promising therapeutic agent for Parkinson disease. As such, there has been great interest in studying its mode of action, which remains unknown. The three-dimensional crystal structure of the N terminus (residues 9-107) of CDNF has been determined, but there have been no published structural studies on the full-length protein due to proteolysis of its C-terminal domain, which is considered intrinsically disordered. An improved purification protocol enabled us to obtain active full-length CDNF and to determine its three-dimensional structure in solution. CDNF contains two well folded domains (residues 10-100 and 111-157) that are linked by a loop of intermediate flexibility. We identified two surface patches on the N-terminal domain that were characterized by increased conformational dynamics that should allow them to embrace active sites. One of these patches is formed by residues Ser-33, Leu-34, Ala-66, Lys-68, Ile-69, Leu-70, Ser-71, and Glu-72. The other includes a flexibly disordered N-terminal tail (residues 1-9), followed by the N-terminal portion of α-helix 1 (residues Cys-11, Glu-12, Val-13, Lys-15, and Glu-16) and residue Glu-88. The surface of the C-terminal domain contains two conserved active sites, which have previously been identified in mesencephalic astrocyte-derived neurotrophic factor, a CDNF paralog, which corresponds to its intracellular mode of action. We also showed that CDNF was able to protect dopaminergic neurons against injury caused by α-synuclein oligomers. This advises its use against physiological damages caused by α-synuclein oligomers, as observed in Parkinson disease and several other neurodegenerative diseases.
- From the Instituto de Bioquímica Médica Leopoldo de Meis, Universidade Federal do Rio de Janeiro, Rio de Janeiro 21.941-902, Brazil.
Organizational Affiliation: