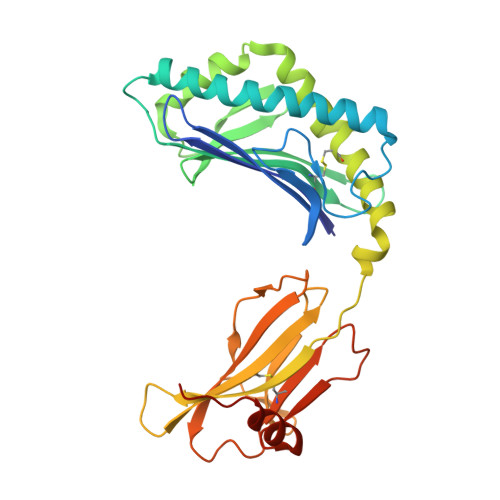
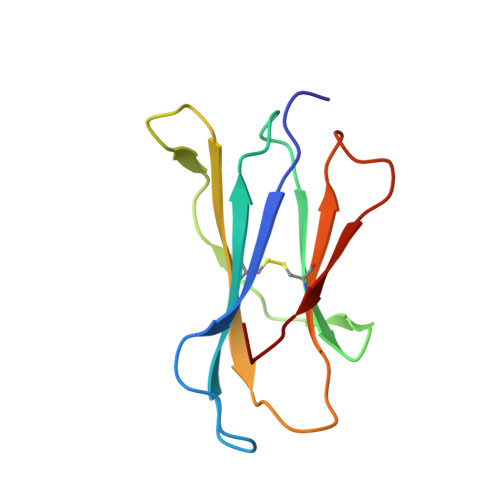
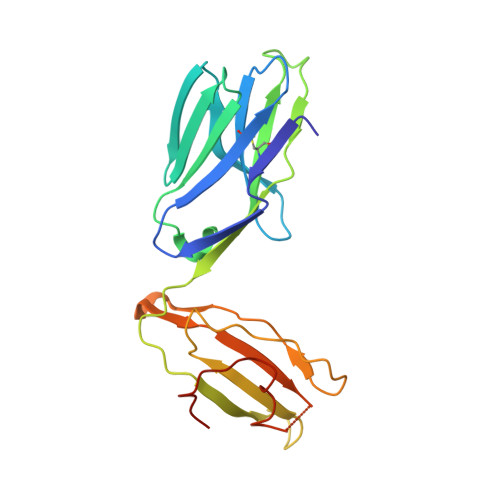
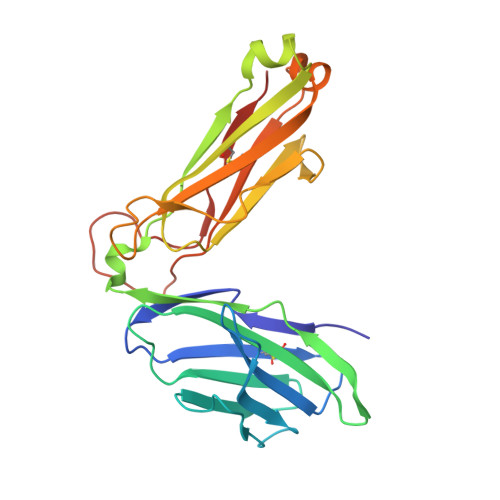
Type-II Natural Killer T Cell Antigen Receptor Mediated Recognition of Cd1D-Sulfatide
Patel, O., Pellicci, D.G., Gras, S., Clarke, A.J., Sandoval-Romero, M., Le Nours, J., Theodossis, A., Mallevaey, T., Gapin, L., Cardell, S., Godfrey, D.I., Rossjohn, J.To be published.