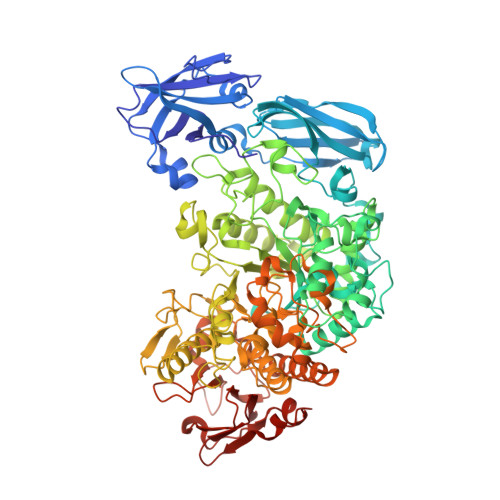
Structure of the Starch-Debranching Enzyme Barley Limit Dextrinase Reveals Homology of the N-Terminal Domain to Cbm21.
Moeller, M.S., Abou Hachem, M., Svensson, B., Henriksen, A.(2012) Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 1008
- PubMed: 22949184
- DOI: https://doi.org/10.1107/S1744309112031004
- Primary Citation of Related Structures:
4AIO - PubMed Abstract:
Barley limit dextrinase (HvLD) is a debranching enzyme from glycoside hydrolase family 13 subfamily 13 (GH13_13) that hydrolyses α-1,6-glucosidic linkages in limit dextrins derived from amylopectin. The structure of HvLD was solved and refined to 1.9 Å resolution. The structure has a glycerol molecule in the active site and is virtually identical to the structures of HvLD in complex with the competitive inhibitors α-cyclodextrin and β-cyclodextrin solved to 2.5 and 2.1 Å resolution, respectively. However, three loops in the N-terminal domain that are shown here to resemble carbohydrate-binding module family 21 were traceable and were included in the present HvLD structure but were too flexible to be traced and included in the structures of the two HvLD-inhibitor complexes.
- Enzyme and Protein Chemistry, Department of Systems Biology, Technical University of Denmark, Søltofts Plads, Building 224, 2800 Kgs. Lyngby, Denmark.
Organizational Affiliation: