Crystal Structure of Inhibitor of Growth 4 (Ing4) Dimerization Domain Reveals Functional Organization of Ing Family of Chromatin-Binding Proteins.
Culurgioni, S., Munoz, I.G., Moreno, A., Palacios, A., Villate, M., Palmero, I., Montoya, G., Blanco, F.J.(2012) J Biological Chem 287: 10876
- PubMed: 22334692
- DOI: https://doi.org/10.1074/jbc.M111.330001
- Primary Citation of Related Structures:
4AFL - PubMed Abstract:
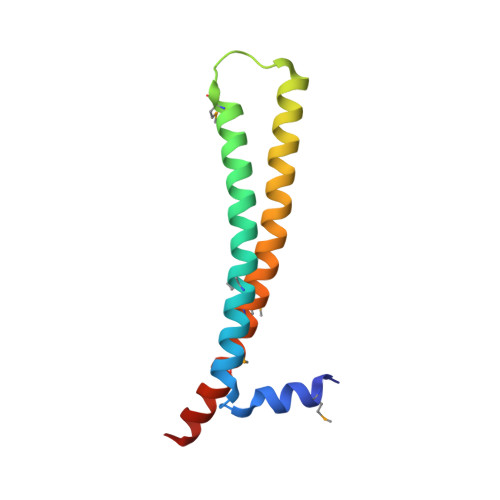
The protein ING4 binds to histone H3 trimethylated at Lys-4 (H3K4me3) through its C-terminal plant homeodomain, thus recruiting the HBO1 histone acetyltransferase complex to target promoters. The structure of the plant homeodomain finger bound to an H3K4me3 peptide has been described, as well as the disorder and flexibility in the ING4 central region. We report the crystal structure of the ING4 N-terminal domain, which shows an antiparallel coiled-coil homodimer with each protomer folded into a helix-loop-helix structure. This arrangement suggests that ING4 can bind simultaneously two histone tails on the same or different nucleosomes. Dimerization has a direct impact on ING4 tumor suppressor activity because monomeric mutants lose the ability to induce apoptosis after genotoxic stress. Homology modeling based on the ING4 structure suggests that other ING dimers may also exist.
- Structural Biology Unit, CIC bioGUNE, E-48160 Derio, Spain.
Organizational Affiliation: