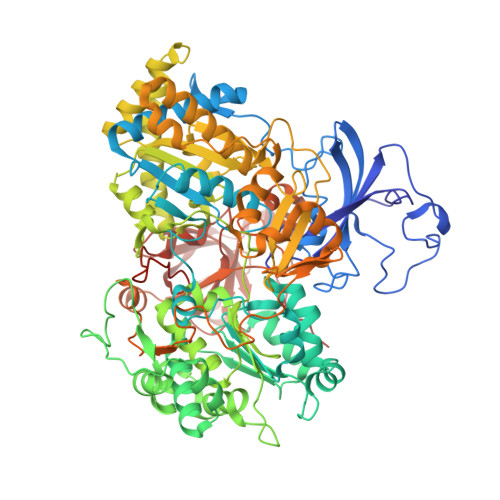
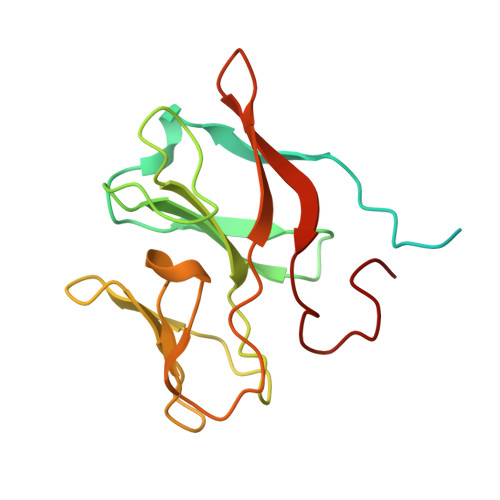
The Respiratory Arsenite Oxidase: Structure and the Role of Residues Surrounding the Rieske Cluster.
Warelow, T.P., Oke, M., Schoepp-Cothenet, B., Dahl, J.U., Bruselat, N., Sivalingam, G.N., Leimkuhler, S., Thalassinos, K., Kappler, U., Naismith, J.H., Santini, J.M.(2013) PLoS One 8: 72535
- PubMed: 24023621
- DOI: https://doi.org/10.1371/journal.pone.0072535
- Primary Citation of Related Structures:
4AAY - PubMed Abstract:
The arsenite oxidase (Aio) from the facultative autotrophic Alphaproteobacterium Rhizobium sp. NT-26 is a bioenergetic enzyme involved in the oxidation of arsenite to arsenate. The enzyme from the distantly related heterotroph, Alcaligenes faecalis, which is thought to oxidise arsenite for detoxification, consists of a large α subunit (AioA) with bis-molybdopterin guanine dinucleotide at its active site and a 3Fe-4S cluster, and a small β subunit (AioB) which contains a Rieske 2Fe-2S cluster. The successful heterologous expression of the NT-26 Aio in Escherichia coli has resulted in the solution of its crystal structure. The NT-26 Aio, a heterotetramer, shares high overall similarity to the heterodimeric arsenite oxidase from A. faecalis but there are striking differences in the structure surrounding the Rieske 2Fe-2S cluster which we demonstrate explains the difference in the observed redox potentials (+225 mV vs. +130/160 mV, respectively). A combination of site-directed mutagenesis and electron paramagnetic resonance was used to explore the differences observed in the structure and redox properties of the Rieske cluster. In the NT-26 AioB the substitution of a serine (S126 in NT-26) for a threonine as in the A. faecalis AioB explains a -20 mV decrease in redox potential. The disulphide bridge in the A. faecalis AioB which is conserved in other betaproteobacterial AioB subunits and the Rieske subunit of the cytochrome bc 1 complex is absent in the NT-26 AioB subunit. The introduction of a disulphide bridge had no effect on Aio activity or protein stability but resulted in a decrease in the redox potential of the cluster. These results are in conflict with previous data on the betaproteobacterial AioB subunit and the Rieske of the bc 1 complex where removal of the disulphide bridge had no effect on the redox potential of the former but a decrease in cluster stability was observed in the latter.
- Institute of Structural and Molecular Biology, University College London, London, United Kingdom.
Organizational Affiliation: