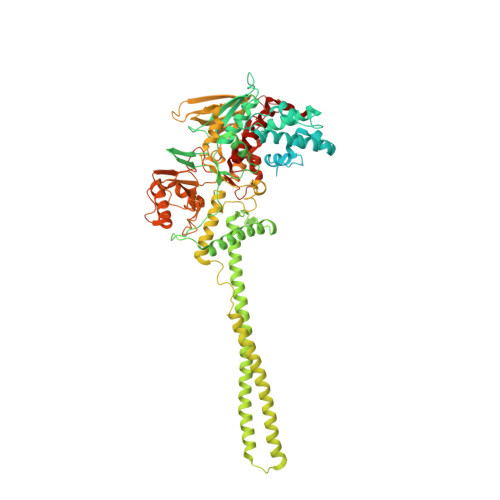
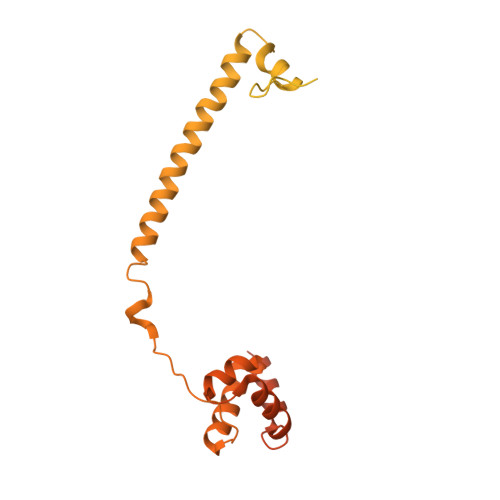
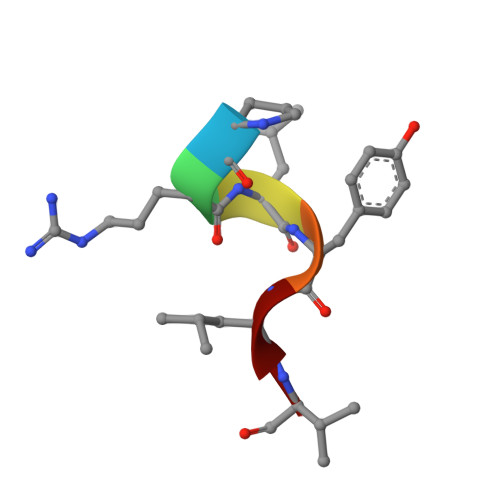
Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Tortorici, M., Borrello, M.T., Tardugno, M., Chiarelli, L.R., Pilotto, S., Ciossani, G., Vellore, N.A., Bailey, S.G., Cowan, J., O'Connell, M., Crabb, S.J., Packham, G.K., Mai, A., Baron, R., Ganesan, A., Mattevi, A.(2013) ACS Chem Biol 8: 1677-1682
- PubMed: 23721412
- DOI: https://doi.org/10.1021/cb4001926
- Primary Citation of Related Structures:
3ZMS, 3ZMT, 3ZMU, 3ZMV, 3ZMZ, 3ZN0, 3ZN1 - PubMed Abstract:
The combinatorial assembly of protein complexes is at the heart of chromatin biology. Lysine demethylase LSD1(KDM1A)/CoREST beautifully exemplifies this concept. The active site of the enzyme tightly associates to the N-terminal domain of transcription factors of the SNAIL1 family, which therefore can competitively inhibit the binding of the N-terminal tail of the histone substrate. Our enzymatic, crystallographic, spectroscopic, and computational studies reveal that LSD1/CoREST can bind to a hexapeptide derived from the SNAIL sequence through recognition of a positively charged α-helical turn that forms upon binding to the enzyme. Variations in sequence and length of this six amino acid ligand modulate affinities enabling the same binding site to differentially interact with proteins that exert distinct biological functions. The discovered short peptide inhibitors exhibit antiproliferative activities and lay the foundation for the development of peptidomimetic small molecule inhibitors of LSD1.
- Department of Biology and Biotechnology, University of Pavia, via Ferrata 9, 27100 Pavia, Italy.
Organizational Affiliation: