Structure of Mutant of Hamp-Dhp Fusion
Zeth, K., Muench, C., Ferris, H.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| HAMP, OSMOLARITY SENSOR PROTEIN ENVZ | 114 | Archaeoglobus fulgidus, Escherichia coli K-12 This entity is chimeric | Mutation(s): 1 EC: 2.7.13.3 | 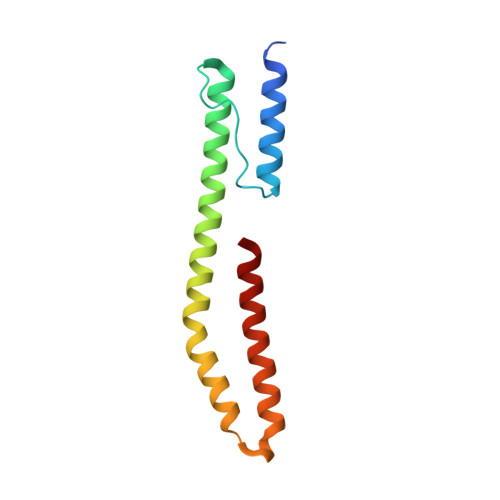 | |
UniProt | |||||
Find proteins for O28769 (Archaeoglobus fulgidus (strain ATCC 49558 / DSM 4304 / JCM 9628 / NBRC 100126 / VC-16)) Explore O28769 Go to UniProtKB: O28769 | |||||
Find proteins for P0AEJ4 (Escherichia coli (strain K12)) Explore P0AEJ4 Go to UniProtKB: P0AEJ4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | P0AEJ4O28769 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 36.06 | α = 90 |
| b = 57.46 | β = 107.84 |
| c = 52.34 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| MOLREP | phasing |