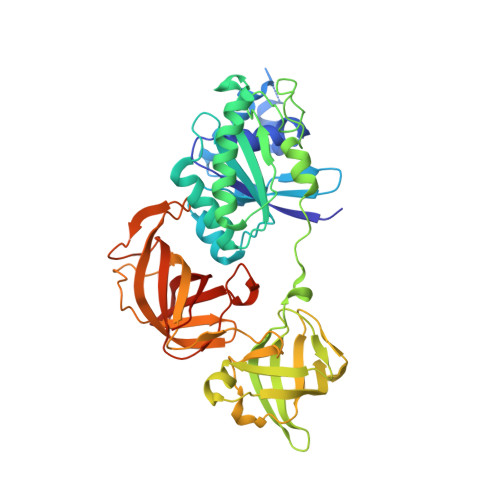
Molecular insights into the interaction of the ribosomal stalk protein with elongation factor 1 alpha.
Ito, K., Honda, T., Suzuki, T., Miyoshi, T., Murakami, R., Yao, M., Uchiumi, T.(2014) Nucleic Acids Res 42: 14042-14052
- PubMed: 25428348
- DOI: https://doi.org/10.1093/nar/gku1248
- Primary Citation of Related Structures:
3WY9, 3WYA - PubMed Abstract:
In all organisms, the large ribosomal subunit contains multiple copies of a flexible protein, the so-called 'stalk'. The C-terminal domain (CTD) of the stalk interacts directly with the translational GTPase factors, and this interaction is required for factor-dependent activity on the ribosome. Here we have determined the structure of a complex of the CTD of the archaeal stalk protein aP1 and the GDP-bound archaeal elongation factor aEF1α at 2.3 Å resolution. The structure showed that the CTD of aP1 formed a long extended α-helix, which bound to a cleft between domains 1 and 3 of aEF1α, and bridged these domains. This binding between the CTD of aP1 and the aEF1α•GDP complex was formed mainly by hydrophobic interactions. The docking analysis showed that the CTD of aP1 can bind to aEF1α•GDP located on the ribosome. An additional biochemical assay demonstrated that the CTD of aP1 also bound to the aEF1α•GTP•aminoacyl-tRNA complex. These results suggest that the CTD of aP1 interacts with aEF1α at various stages in translation. Furthermore, phylogenetic perspectives and functional analyses suggested that the eukaryotic stalk protein also interacts directly with domains 1 and 3 of eEF1α, in a manner similar to the interaction of archaeal aP1 with aEF1α.
- Department of Biology, Faculty of Science, Niigata University, 8050 Ikarashi 2-no-cho, Nishi-ku, Niigata 950-2181, Japan uchiumi@bio.sc.niigata-u.ac.jp.
Organizational Affiliation: