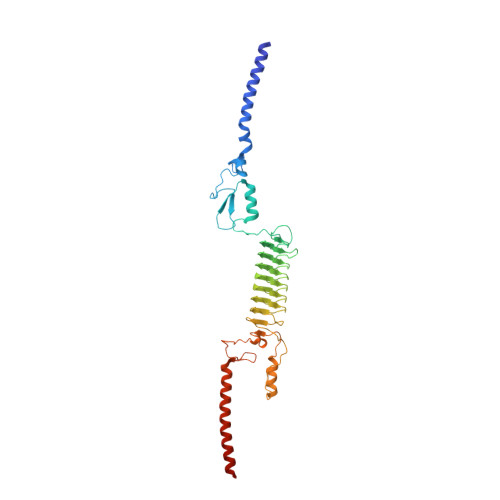
Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
Koiwai, K., Hartmann, M.D., Linke, D., Lupas, A.N., Hori, K.(2016) J Biological Chem 291: 3705-3724
- PubMed: 26698633
- DOI: https://doi.org/10.1074/jbc.M115.701698
- Primary Citation of Related Structures:
3WP8, 3WPA, 3WPO, 3WPP, 3WPR, 3WQA - PubMed Abstract:
Trimeric autotransporter adhesins (TAAs) on the cell surface of Gram-negative pathogens mediate bacterial adhesion to host cells and extracellular matrix proteins. However, AtaA, a TAA in the nonpathogenic Acinetobacter sp. strain Tol 5, shows nonspecific high adhesiveness to abiotic material surfaces as well as to biotic surfaces. It consists of a passenger domain secreted by the C-terminal transmembrane anchor domain (TM), and the passenger domain contains an N-terminal head, N-terminal stalk, C-terminal head (Chead), and C-terminal stalk (Cstalk). The Chead-Cstalk-TM fragment, which is conserved in many Acinetobacter TAAs, has by itself the head-stalk-anchor architecture of a complete TAA. Here, we show the crystal structure of the Chead-Cstalk fragment, AtaA_C-terminal passenger domain (CPSD), providing the first view of several conserved TAA domains. The YadA-like head (Ylhead) of the fragment is capped by a unique structure (headCap), composed of three β-hairpins and a connector motif; it also contains a head insert motif (HIM1) before its last inner β-strand. The headCap, Ylhead, and HIM1 integrally form a stable Chead structure. Some of the major domains of the CPSD fragment are inherently flexible and provide bending sites for the fiber between segments whose toughness is ensured by topological chain exchange and hydrophobic core formation inside the trimer. Thus, although adherence assays using in-frame deletion mutants revealed that the characteristic adhesive sites of AtaA reside in its N-terminal part, the flexibility and toughness of the CPSD part provide the resilience that enables the adhesive properties of the full-length fiber across a wide range of conditions.
- From the Department of Biotechnology, Graduate School of Engineering, Nagoya University, Chikusa-ku, Nagoya 464-8603, Japan, the Structural Biology Research Center, Photon Factory, Institute of Materials Structure Science, High Energy Accelerator Research Organization, Tsukuba 305-0801, Japan.
Organizational Affiliation: