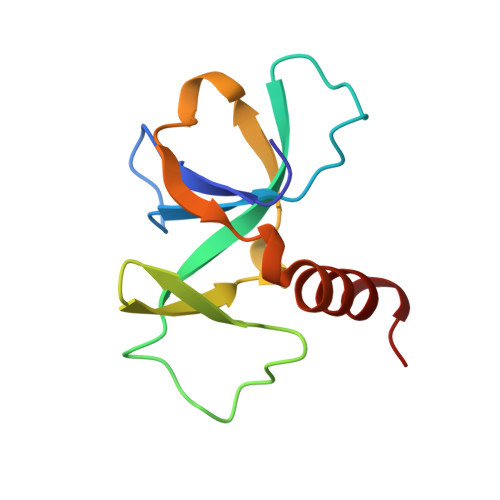
Crystal structure of CcdB, a topoisomerase poison from E. coli.
Loris, R., Dao-Thi, M.-H., Bahassi, E.M., Van Melderen, L., Poortmans, F., Liddington, R., Couturier, M., Wyns, L.(1999) J Mol Biology 285: 1667-1677
- PubMed: 9917404
- DOI: https://doi.org/10.1006/jmbi.1998.2395
- Primary Citation of Related Structures:
1VUB, 2VUB, 3VUB, 4VUB - PubMed Abstract:
The crystal structure of CcdB, a protein that poisons Escherichia coli gyrase, was determined in three crystal forms. The protein consists of a five-stranded antiparallel beta-pleated sheet followed by a C-terminal alpha-helix. In one of the loops of the sheet, a second small three-stranded antiparallel beta-sheet is inserted that sticks out of the molecule as a wing. This wing contains the LysC proteolytic cleavage site that is protected by CcdA and, therefore, forms a likely CcdA recognition site. A dimer is formed by sheet extension and by extensive hydrophobic contacts involving three of the five methionine residues and the C terminus of the alpha-helix. The surface of the dimer on the side of the alpha-helix is overall negatively charged, while the opposite side as well as the wing sheet is dominated by positive charges. We propose that the CcdB dimer binds into the central hole of the 59 kDa N-terminal fragment of GyrA, after disruption of the head dimer interface of GyrA.
- Laboratorium voor Ultrastructuur, Vlaams Interuniversitair Instituut voor Biotechnologie, Vrije Universiteit Brussel, Paardenstraat 65, Sint-Genesius-Rode, B-1640, Belgium. reloris@vub.ac.be
Organizational Affiliation: