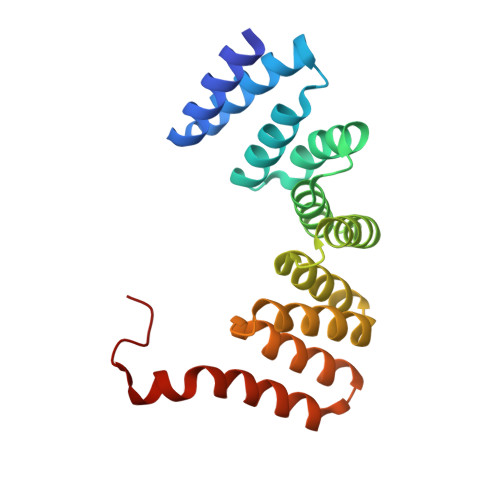
Inter-phylum structural conservation of the magnetosome-associated TPR-containing protein, MamA
Zeytuni, N., Baran, D., Davidov, G., Zarivach, R.(2012) J Struct Biol 180: 479-487
- PubMed: 22917855
- DOI: https://doi.org/10.1016/j.jsb.2012.08.001
- Primary Citation of Related Structures:
3VTX, 3VTY - PubMed Abstract:
Magnetotactic bacteria enclose the magnetosome, a unique prokaryotic sub-cellular organelle that allows the biomineralization of magnetic nano-crystals. Membrane-coated magnetosomes are arranged into a linear chain that permits magnetotactic bacteria to navigate geomagnetic fields. Magnetosome assembly and biomineralization are controlled by conserved magnetosome-associated proteins, including MamA, a tetra-trico-peptide repeat (TPR)-containing protein that was shown to coat the magnetosome membrane. In this study, two MamA structures from Candidatus Magnetobacterium bavaricum (Mbav) were determined via X-ray crystallography. These structures confirm that Mbav MamA folds as a sequential TPR protein and shares a high degree of structural similarity with homologous MamA proteins from Magnetospirillum species. Furthermore, the two TPR-containing domains of MamA are separated by an interphylum-conserved region containing a flexible hinge that is involved in ligand binding and recognition. Finally, substantial differences were found in the local stabilization of the MamA N-terminal domain as a result of the loss of an evolutionary conserved salt bridge.
- Department of Life Sciences, Ben Gurion University of the Negev, P.O. Box 653, Beer-Sheva 84105, Israel.
Organizational Affiliation: