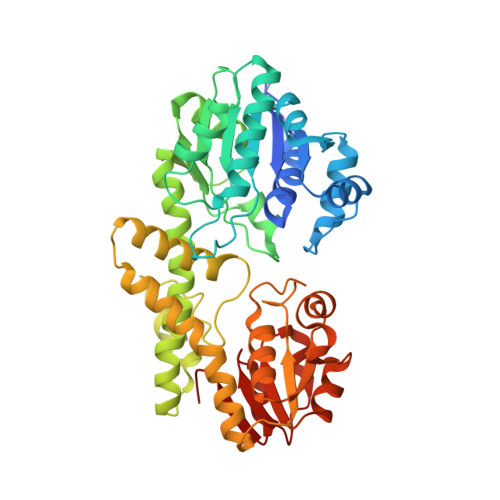
Structure of a UDP-glucose dehydrogenase from the hyperthermophilic archaeon Pyrobaculum islandicum.
Sakuraba, H., Kawai, T., Yoneda, K., Ohshima, T.(2012) Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 1003-1007
- PubMed: 22949183
- DOI: https://doi.org/10.1107/S1744309112030667
- Primary Citation of Related Structures:
3VTF - PubMed Abstract:
The crystal structure of an extremely thermostable UDP-glucose dehydrogenase (UDP-GDH) from the hyperthermophilic archaeon Pyrobaculum islandicum was determined at a resolution of 2.0 Å. The overall fold was comprised of an N-terminal NAD(+) dinucleotide binding domain and a C-terminal UDP-sugar binding domain connected by a long α-helix, and the main-chain coordinates of the enzyme were similar to those of previously studied UDP-GDHs, including the enzymes from Burkholderia cepacia, Streptococcus pyogenes and Klebsiella pneumoniae. However, the sizes of several surface loops in P. islandicum UDP-GDH were much smaller than the corresponding loops in B. cepacia UDP-GDH but were comparable to those of the S. pyogenes and K. pneumoniae enzymes. Structural comparison revealed that the presence of extensive intersubunit hydrophobic interactions, as well as the formation of an intersubunit aromatic pair network, is likely to be the main factor contributing to the hyperthermostability of P. islandicum UDP-GDH.
- Department of Applied Biological Science, Faculty of Agriculture, Kagawa University, 2393 Ikenobe, Kita-gun, Kagawa 761-0795, Japan.
Organizational Affiliation: