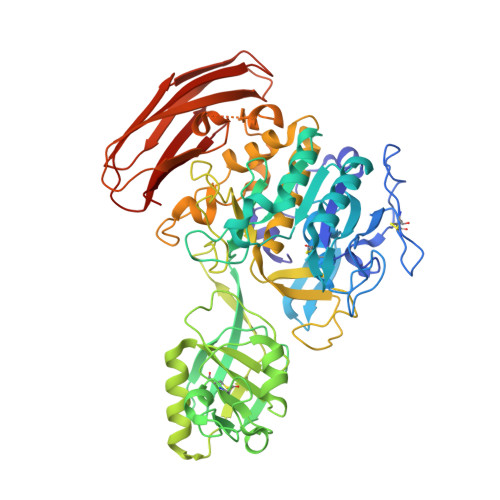
Crystal structure of cucumisin, a subtilisin-like endoprotease from Cucumis melo L
Murayama, K., Kato-Murayama, M., Hosaka, T., Sotokawauchi, A., Yokoyama, S., Arima, K., Shirouzu, M.(2012) J Mol Biology 423: 386-396
- PubMed: 22841692
- DOI: https://doi.org/10.1016/j.jmb.2012.07.013
- Primary Citation of Related Structures:
3VTA - PubMed Abstract:
Cucumisin is a plant serine protease, isolated as an extracellular glycoprotein from the melon fruit Cucumis melo L. var. Prince. Cucumisin is composed of multiple domain modules, including catalytic, protease-associated, and fibronectin-III-like domains. The crystal structure of cucumisin was determined by the multiwavelength anomalous dispersion method and refined at 2.75Å resolution. A structural homology search indicated that the catalytic domain of cucumisin shares structural similarity with subtilisin and subtilisin-like fold enzymes. According to the Z-score, the highest structural similarity is with tomato subtilase 3 (SBT3), with an rmsd of 3.5Å for the entire region. The dimer formation mediated by the protease-associated domain in SBT3 is a distinctive structural characteristic of cucumisin. On the other hand, analytical ultracentrifugation indicated that cucumisin is mainly monomeric in solution. Although the locations of the amino acid residues composing the catalytic triad are well conserved between cucumisin and SBT3, a disulfide bond is uniquely located near the active site of cucumisin. The steric circumstances of the active site with this disulfide bond are distinct from those of SBT3, and it contributes to the substrate preference of cucumisin, especially at the P2 position. Among the plant serine proteases, the thermostability of cucumisin is higher than that of its structural homologue SBT3, as determined by their melting points. A structural comparison between cucumisin and SBT3 revealed that cucumisin possesses less surface area and shortened loop regions. Consequently, the higher thermostability of cucumisin is achieved by its more compact structure.
- Division of Biomedical Measurements and Diagnostics, Graduate School of Biomedical Engineering, Tohoku University, Sendai 980-8575, Japan.
Organizational Affiliation: