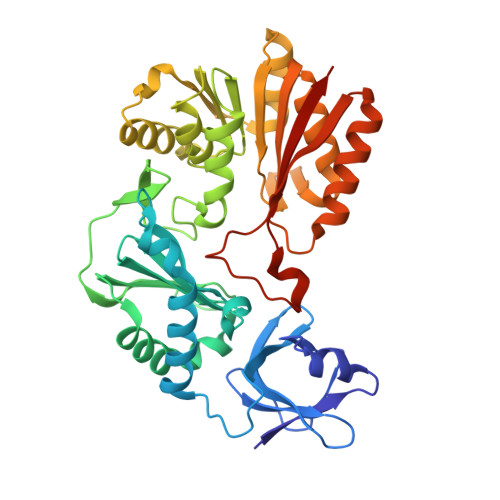
Crystal structure of a putative methyltransferase SAV1081 from Staphylococcus aureus
Kita, S., Tanaka, Y., Hirano, N., Kimura, S., Suzuki, T., Suzuki, T., Yao, M., Tanaka, I.(2012) Protein Pept Lett 20: 530-537
- PubMed: 23016631
- DOI: https://doi.org/10.2174/0929866511320050006
- Primary Citation of Related Structures:
3VSE - PubMed Abstract:
Cluster of Orthologous Groups (COG) 1092 contains two distinct types of methylation enzyme from Escherichia coli, YccW and YcbY. YccW is a 5-methylcytosine methyltransferase (m5C MTase) responsible for m5C 1962 in 23S rRNA, whereas YcbY is a dimethyltransferase, of which N- and C-terminal domains are responsible for N2- methylguanosine (m2G) 2445 and 7-methylguanosine (m7G) 2069 in 23S rRNA, respectively. However, proteins in COG1092 other than YccW and YcbY remain functionally unidentified. SAV1081 from Staphylococcus aureus is one of the functionally unassigned proteins of COG1092. Although SAV1081 has an identical domain organization to YccW with 26% sequence identity, it lacks the catalytic cysteine residue essential for m5C formation activity. In the present study, we determined the crystal structure of SAV1081 and compared it with those of other COG1092 proteins. Based on the structure characteristics, such as the presence or absence of the catalytic cysteine residue, β-hairpin structure, and oligomeric state, as well as domain organization, we propose a functional classification of COG1092 proteins.
- Graduate School of Life Sciences, Hokkaido University, Sapporo, 060-0810, Japan.
Organizational Affiliation: