Crystal structure of mitochondrial quinol-fumarate reductase from parasitic nematode Ascaris suum
Shimizu, H., Shiba, T., Inaoka, D.K., Osanai, A., Kita, K., Sakamoto, K., Harada, S.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Flavoprotein subunit of complex II | 645 | Ascaris suum | Mutation(s): 0 EC: 1.3.5.1 Membrane Entity: Yes | 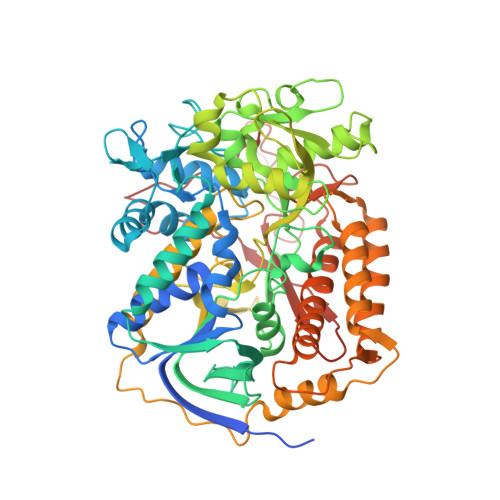 | |
UniProt | |||||
Find proteins for Q33862 (Ascaris suum) Explore Q33862 Go to UniProtKB: Q33862 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q33862 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Iron-sulfur subunit of succinate dehydrogenase | 282 | Ascaris suum | Mutation(s): 0 EC: 1.3.5.1 Membrane Entity: Yes | 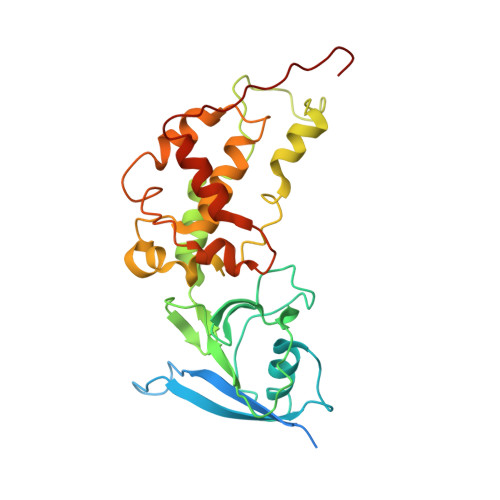 | |
UniProt | |||||
Find proteins for O44074 (Ascaris suum) Explore O44074 Go to UniProtKB: O44074 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O44074 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome b-large subunit | 188 | Ascaris suum | Mutation(s): 0 Membrane Entity: Yes | 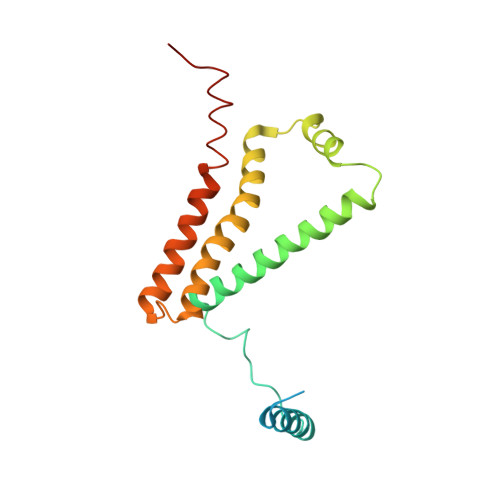 | |
UniProt | |||||
Find proteins for P92506 (Ascaris suum) Explore P92506 Go to UniProtKB: P92506 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P92506 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Succinate dehydrogenase [ubiquinone] cytochrome b small subunit, mitochondrial | 156 | Ascaris suum | Mutation(s): 0 Membrane Entity: Yes | 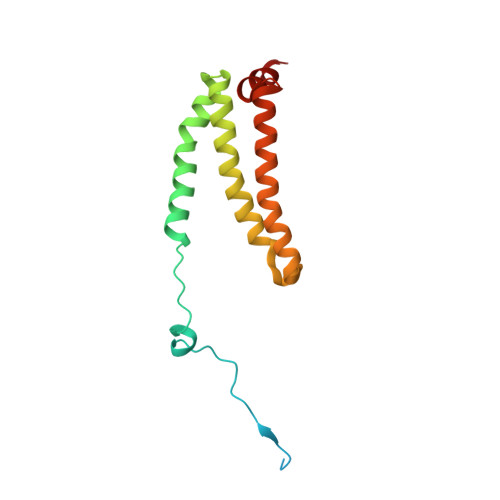 | |
UniProt | |||||
Find proteins for P92507 (Ascaris suum) Explore P92507 Go to UniProtKB: P92507 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P92507 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 8 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FAD Query on FAD | J [auth A], R [auth E] | FLAVIN-ADENINE DINUCLEOTIDE C27 H33 N9 O15 P2 VWWQXMAJTJZDQX-UYBVJOGSSA-N |  | ||
| EPH Query on EPH | P [auth D], X [auth H] | L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE C39 H68 N O8 P MABRTXOVHMDVAT-AAEGOEIASA-N |  | ||
| HEM Query on HEM | N [auth C], W [auth G] | PROTOPORPHYRIN IX CONTAINING FE C34 H32 Fe N4 O4 KABFMIBPWCXCRK-RGGAHWMASA-L |  | ||
| SF4 Query on SF4 | L [auth B], T [auth F] | IRON/SULFUR CLUSTER Fe4 S4 LJBDFODJNLIPKO-UHFFFAOYSA-N |  | ||
| FTN Query on FTN | O [auth C], V [auth F] | N-[3-(1-methylethoxy)phenyl]-2-(trifluoromethyl)benzamide C17 H16 F3 N O2 PTCGDEVVHUXTMP-UHFFFAOYSA-N |  | ||
| F3S Query on F3S | M [auth B], U [auth F] | FE3-S4 CLUSTER Fe3 S4 FCXHZBQOKRZXKS-UHFFFAOYSA-N |  | ||
| FES Query on FES | K [auth B], S [auth F] | FE2/S2 (INORGANIC) CLUSTER Fe2 S2 NIXDOXVAJZFRNF-UHFFFAOYSA-N |  | ||
| MLI Query on MLI | I [auth A], Q [auth E] | MALONATE ION C3 H2 O4 OFOBLEOULBTSOW-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 124.011 | α = 90 |
| b = 135.333 | β = 90 |
| c = 220.498 | γ = 90 |
| Software Name | Purpose |
|---|---|
| MOLREP | phasing |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |