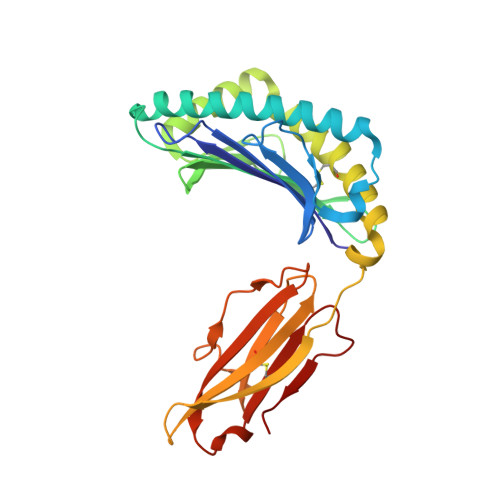
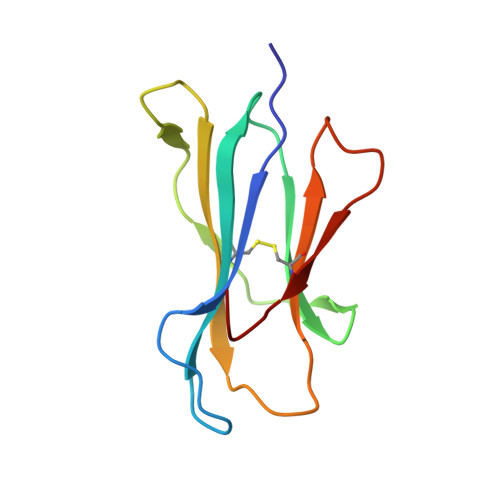
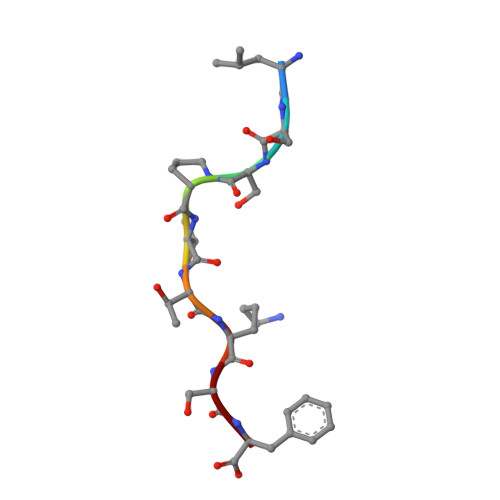
Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B
Vivian, J.P., Duncan, R.C., Berry, R., O'Connor, G.M., Reid, H.H., Beddoe, T., Gras, S., Saunders, P.M., Olshina, M.A., Widjaja, J.M.L., Harpur, C.M., Lin, J., Maloveste, S.M., Price, D.A., Lafont, B.A.P., McVicar, D.W., Clements, C.S., Brooks, A.G., Rossjohn, J.(2011) Nature 479: 401-405
- PubMed: 22020283
- DOI: https://doi.org/10.1038/nature10517
- Primary Citation of Related Structures:
3VH8 - PubMed Abstract:
Members of the killer cell immunoglobulin-like receptor (KIR) family, a large group of polymorphic receptors expressed on natural killer (NK) cells, recognize particular peptide-laden human leukocyte antigen (pHLA) class I molecules and have a pivotal role in innate immune responses. Allelic variation and extensive polymorphism within the three-domain KIR family (KIR3D, domains D0-D1-D2) affects pHLA binding specificity and is linked to the control of viral replication and the treatment outcome of certain haematological malignancies. Here we describe the structure of a human KIR3DL1 receptor bound to HLA-B*5701 complexed with a self-peptide. KIR3DL1 clamped around the carboxy-terminal end of the HLA-B*5701 antigen-binding cleft, resulting in two discontinuous footprints on the pHLA. First, the D0 domain, a distinguishing feature of the KIR3D family, extended towards β2-microglobulin and abutted a region of the HLA molecule with limited polymorphism, thereby acting as an 'innate HLA sensor' domain. Second, whereas the D2-HLA-B*5701 interface exhibited a high degree of complementarity, the D1-pHLA-B*5701 contacts were suboptimal and accommodated a degree of sequence variation both within the peptide and the polymorphic region of the HLA molecule. Although the two-domain KIR (KIR2D) and KIR3DL1 docked similarly onto HLA-C and HLA-B respectively, the corresponding D1-mediated interactions differed markedly, thereby providing insight into the specificity of KIR3DL1 for discrete HLA-A and HLA-B allotypes. Collectively, in association with extensive mutagenesis studies at the KIR3DL1-pHLA-B*5701 interface, we provide a framework for understanding the intricate interplay between peptide variability, KIR3D and HLA polymorphism in determining the specificity requirements of this essential innate interaction that is conserved across primate species.
- Department of Biochemistry and Molecular Biology, School of Biomedical Sciences, Monash University, Clayton, Victoria 3800, Australia.
Organizational Affiliation: