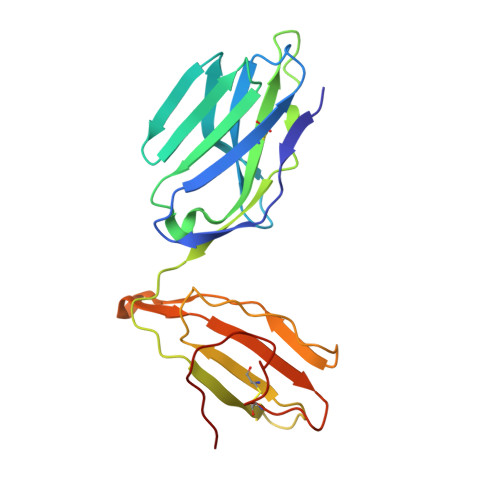
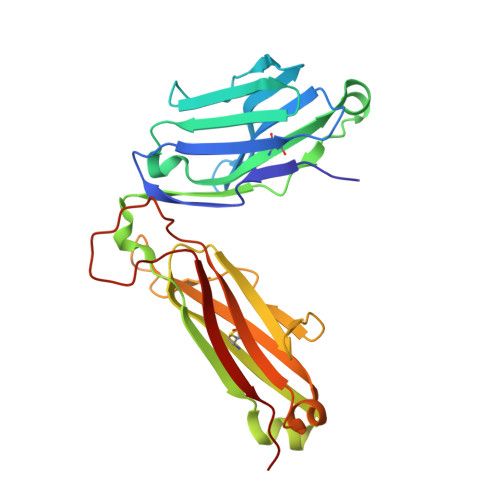
Structural basis for the killing of human beta cells by CD8(+) T cells in type 1 diabetes.
Bulek, A.M., Cole, D.K., Skowera, A., Dolton, G., Gras, S., Madura, F., Fuller, A., Miles, J.J., Gostick, E., Price, D.A., Drijfhout, J.W., Knight, R.R., Huang, G.C., Lissin, N., Molloy, P.E., Wooldridge, L., Jakobsen, B.K., Rossjohn, J., Peakman, M., Rizkallah, P.J., Sewell, A.K.(2012) Nat Immunol 13: 283-289
- PubMed: 22245737
- DOI: https://doi.org/10.1038/ni.2206
- Primary Citation of Related Structures:
3UTP, 3UTQ, 3UTS, 3UTT - PubMed Abstract:
The structural characteristics of the engagement of major histocompatibility complex (MHC) class II-restricted self antigens by autoreactive T cell antigen receptors (TCRs) is established, but how autoimmune TCRs interact with complexes of self peptide and MHC class I has been unclear. Here we examined how CD8(+) T cells kill human islet beta cells in type 1 diabetes via recognition of a human leukocyte antigen HLA-A*0201-restricted glucose-sensitive preproinsulin peptide by the autoreactive TCR 1E6. Rigid 'lock-and-key' binding underpinned the 1E6-HLA-A*0201-peptide interaction, whereby 1E6 docked similarly to most MHC class I-restricted TCRs. However, this interaction was extraordinarily weak because of limited contacts with MHC class I. TCR binding was highly peptide centric, dominated by two residues of the complementarity-determining region 3 (CDR3) loops that acted as an 'aromatic-cap' over the complex of peptide and MHC class I (pMHCI). Thus, highly focused peptide-centric interactions associated with suboptimal TCR-pMHCI binding affinities might lead to thymic escape and potential CD8(+) T cell-mediated autoreactivity.
- Institute of Infection and Immunity, Cardiff University School of Medicine, Heath Park, Cardiff, UK.
Organizational Affiliation: