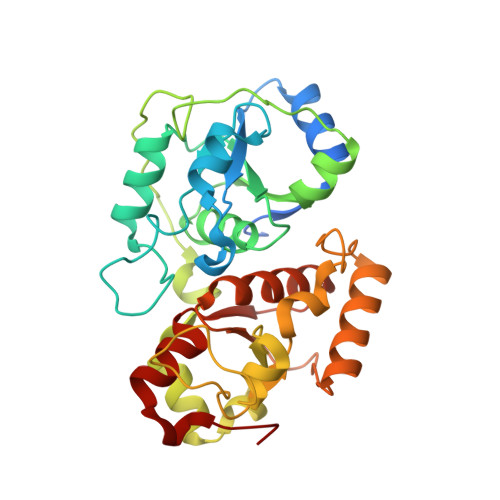
Crystal structure of the Tum1 protein from the yeast Saccharomyces cerevisiae.
Qiu, R., Wang, F., Liu, M., Lou, T., Ji, C.(2012) Protein Pept Lett 19: 1139-1143
- PubMed: 22587783
- DOI: https://doi.org/10.2174/092986612803217060
- Primary Citation of Related Structures:
3UTN - PubMed Abstract:
Yeast tRNA-thiouridine modification protein 1 (Tum1) plays essential role in the sulfur transfer process of Urm1 system, which in turn is involved in many important cellular processes. In the rhodanese-like domain (RLD), conserved cysteine residue is proved to be the centre of active site of sulfurtransferases and crucial for the substrate recognition. In this report, we describe the crystal structure of Tum1 protein at 1.90 A resolution which, despite consisting of two RLDs, has only one conserved cysteine residue in the C-terminal RLD. An unaccounted electron density is found near the active site, which might point to the new cofactor in the sulfur transfer mechanism.
- State Key Laboratory of Genetic Engineering, Institute of Genetics, School of Life Sciences, Fudan University, Shanghai, People’s Republic of China.
Organizational Affiliation: