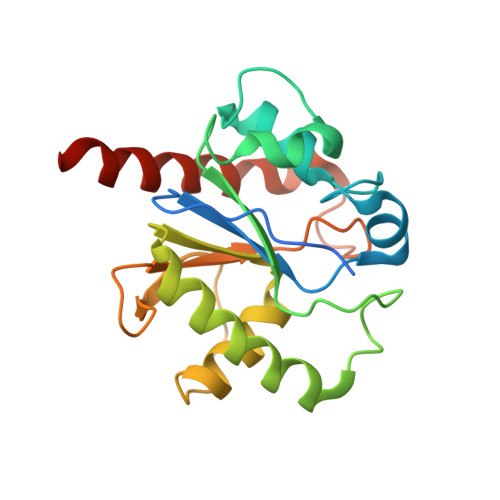
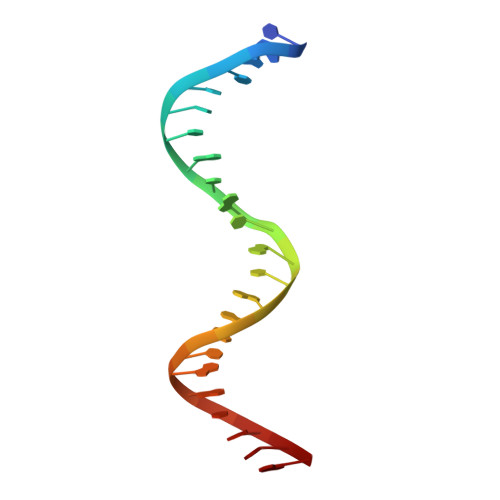
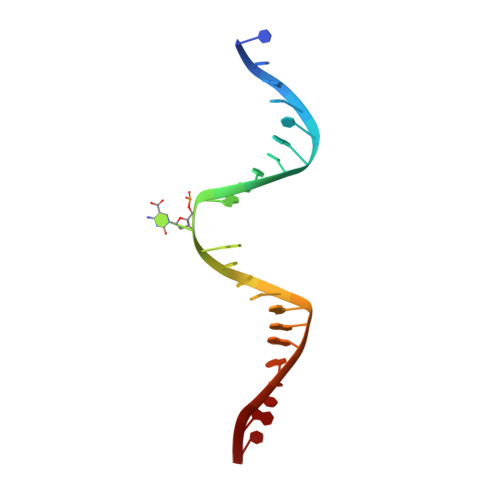
Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA.
Zhang, L., Lu, X., Lu, J., Liang, H., Dai, Q., Xu, G.L., Luo, C., Jiang, H., He, C.(2012) Nat Chem Biol 8: 328-330
- PubMed: 22327402
- DOI: https://doi.org/10.1038/nchembio.914
- Primary Citation of Related Structures:
3UO7, 3UOB - PubMed Abstract:
Human thymine DNA glycosylase (hTDG) efficiently excises 5-carboxylcytosine (5caC), a key oxidation product of 5-methylcytosine in genomic DNA, in a recently discovered cytosine demethylation pathway. We present here the crystal structures of the hTDG catalytic domain in complex with duplex DNA containing either 5caC or a fluorinated analog. These structures, together with biochemical and computational analyses, reveal that 5caC is specifically recognized in the active site of hTDG, supporting the role of TDG in mammalian 5-methylcytosine demethylation.
- Department of Chemistry and Institute for Biophysical Dynamics, The University of Chicago, Chicago, Illinois, USA.
Organizational Affiliation: