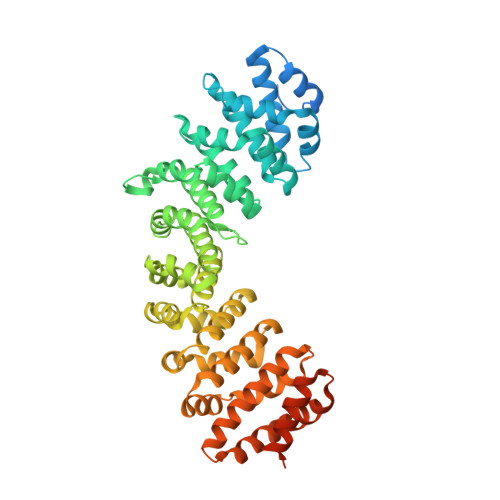
Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-alpha
Marfori, M., Lonhienne, T.G., Forwood, J.K., Kobe, B.(2012) Traffic 13: 532-548
- PubMed: 22248489
- DOI: https://doi.org/10.1111/j.1600-0854.2012.01329.x
- Primary Citation of Related Structures:
3UKW, 3UKX, 3UKY, 3UKZ, 3UL0, 3UL1 - PubMed Abstract:
Classical nuclear localization signals (cNLSs), comprising one (monopartite cNLSs) or two clusters of basic residues connected by a 10-12 residue linker (bipartite cNLSs), are recognized by the nuclear import factor importin-α. The cNLSs bind along a concave groove on importin-α; however, specificity determinants of cNLSs remain poorly understood. We present a structural and interaction analysis study of importin-α binding to both designed and naturally occurring high-affinity cNLS-like sequences; the peptide inhibitors Bimax1 and Bimax2, and cNLS peptides of cap-binding protein 80. Our data suggest that cNLSs and cNLS-like sequences can achieve high affinity through maximizing interactions at the importin-α minor site, and by taking advantage of multiple linker region interactions. Our study defines an extended set of binding cavities on the importin-α surface, and also expands on recent observations that longer linker sequences are allowed, and that long-range electrostatic complementarity can contribute to cNLS-binding affinity. Altogether, our study explains the molecular and structural basis of the results of a number of recent studies, including systematic mutagenesis and peptide library approaches, and provides an improved level of understanding on the specificity determinants of a cNLS. Our results have implications for identifying cNLSs in novel proteins.
- School of Chemistry and Molecular Biosciences, University of Queensland, Brisbane, QLD, 4072, Australia.
Organizational Affiliation: