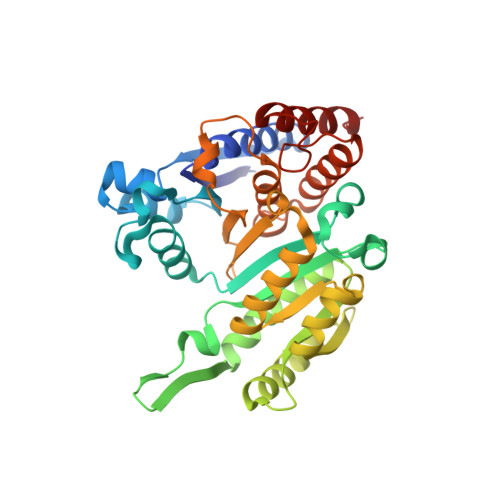
Crystal structure of putative 3-isopropylmalate dehydrogenase from Campylobacter jejuni
Tkaczuk, K.L., Chruszcz, M., Grimshaw, S., Onopriyenko, O., Savchenko, A., Anderson, W.F., Minor, W., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.