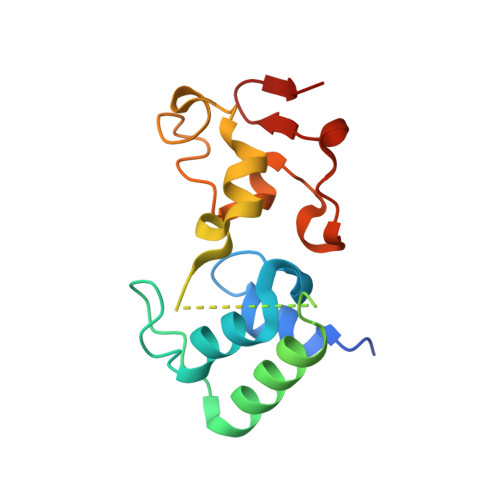
High-resolution crystal structure of the recombinant diheme cytochrome c from Shewanella baltica (OS155).
De March, M., Di Rocco, G., Hickey, N., Geremia, S.(2015) J Biomol Struct Dyn 33: 395-403
- PubMed: 24559494
- DOI: https://doi.org/10.1080/07391102.2014.880657
- Primary Citation of Related Structures:
3U99 - PubMed Abstract:
Multiheme cytochromes c (cyts c) are c-type cyts characterized by non-standard structural and spectroscopic properties. The relative disposition of the heme cofactors in the core of these proteins is conserved and they can be classified from their geometry in two main groups. In one group the porphyrin planes are arranged in a perpendicular fashion, while in the other they are parallel. Orientation of the heme groups is a key factor that regulates the intramolecular electron transfer pathway. A 16.5 kDa diheme cyt c, isolated from the bacterium Shewanella baltica OS155 (Sb-DHC), was cloned and expressed in E. coli and its structure was investigated by X-ray crystallography. Using high-resolution data (1.14 Å) collected at ELETTRA (Trieste), the crystal structure, with an orthorhombic cell (a = 40.81, b = 42.97, c = 82.07 Å), was solved using the homologous diheme from Rhodobacter sphaeroides (Rs-DHC) as the initial model. The electron density map of the refined structure (Rfact of 13.8% and Rfree of 15.4%) shows a two domain structure connected by a central unstructured region (N72-G87). The Sb-DHC, like its homologue (Rs-DHC), folds into a new cyt c class: the N-terminal globular domain, with its three α-helices, belongs to class I of c-type cyts, while the C-terminal domain includes a rare π-helix. The metal centre of the c-type heme groups is axially coordinated by two His residues and it is covalently bound to the protein through two Cys bonds.
- a Centre of Excellence in Biocrystallography (CEB), Department of Chemical and Pharmaceutical Sciences , University of Trieste , via L.Giorgieri 1, 34127 Trieste , Italy.
Organizational Affiliation: