The crystal structure of FMN-dependent NADH-azoreductase 1 (GBAA0966) from Bacillus anthracis str. Ames Ancestor
Zhang, R., Gu, M., Tan, K., Kwon, K., Anderson, W.F., Joachimiak, A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| FMN-dependent NADH-azoreductase 1 | 223 | Bacillus anthracis str. 'Ames Ancestor | Mutation(s): 0 Gene Names: azoR1, Bacillus anthracis, BAS0908, BA_0966, GBAA_0966 EC: 1.7 (PDB Primary Data), 1.6.5 (UniProt), 1.7.1.17 (UniProt) | 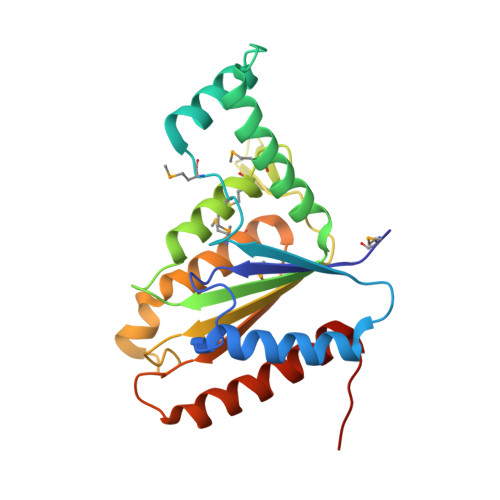 | |
UniProt | |||||
Find proteins for Q81UB2 (Bacillus anthracis) Explore Q81UB2 Go to UniProtKB: Q81UB2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q81UB2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PEG Query on PEG | I [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | G [auth A], H [auth A], M [auth B], P [auth D] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| CL Query on CL | E [auth A] F [auth A] J [auth B] K [auth B] L [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B, C, D | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 102.802 | α = 90 |
| b = 113.47 | β = 90.31 |
| c = 91.115 | γ = 90 |
| Software Name | Purpose |
|---|---|
| SBC-Collect | data collection |
| MOLREP | phasing |
| PHENIX | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |