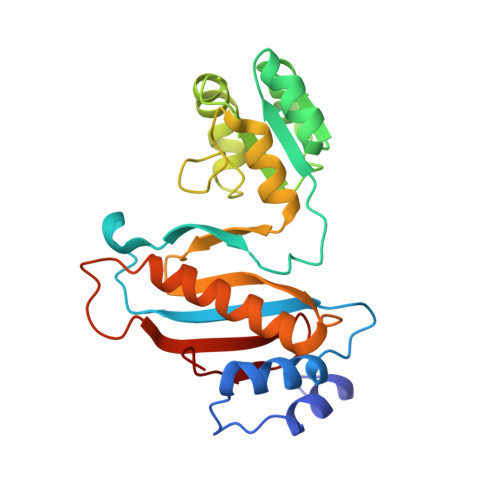
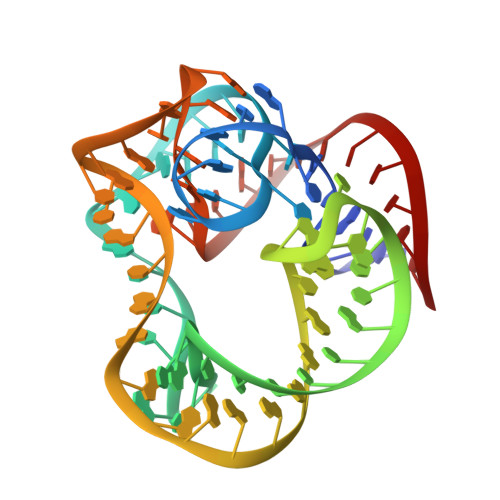
Crystal structure of ribosomal protein tthl1 in complex with 80nt 23s rna from thermus thermophilus
Tishchenko, S.V., Nikonova, E.Y., Kostareva, O.S., Gabdulkhakov, A.G., Sarskikh, A.V., Piendl, W., Nikonov, S.V., Garber, M.B., Nevskaya, N.A.To be published.