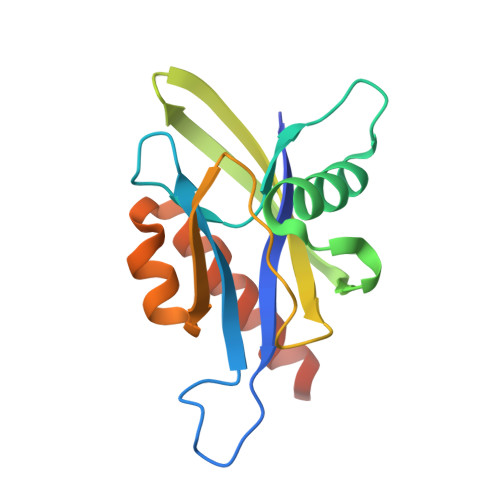
Crystal structure of wild-type and mutant human Ap4A hydrolase
Ge, H.H., Chen, X.F., Yang, W.L., Niu, L.W., Teng, M.K.(2013) Biochem Biophys Res Commun 432: 16-21
- PubMed: 23384440
- DOI: https://doi.org/10.1016/j.bbrc.2013.01.095
- Primary Citation of Related Structures:
3U53, 4ICK, 4IJX - PubMed Abstract:
Ap4A hydrolase (asymmetrical diadenosine tetraphosphate hydrolase, EC 3.6.1.17), an enzyme involved in a number of biological processes, is characterized as cleaving the polyphosphate chain at the fourth phosphate from the bound adenosine moiety. This paper presents the crystal structure of wild-type and E58A mutant human Ap4A hydrolase. Similar to the canonical Nudix fold, human Ap4A hydrolase shows the common αβα-sandwich architecture. Interestingly, two sulfate ions and one diphosphate coordinated with some conserved residues were observed in the active cleft, which affords a better understanding of a possible mode of substrate binding.
- Institute of Health Sciences and Modern Experiment Technology Center, Anhui University, Hefei 230601, People's Republic of China. hhge@ahu.edu.cn
Organizational Affiliation: