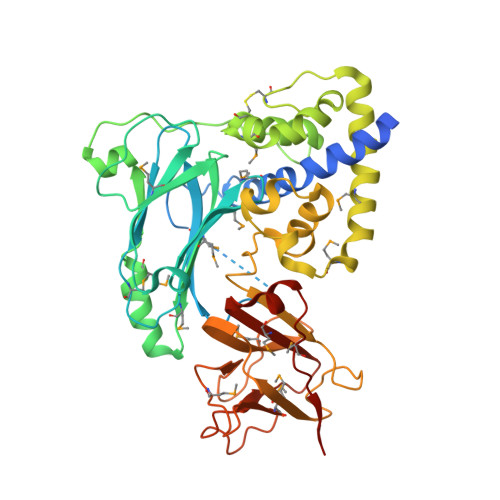
Crystal Structure of the VPA0106 protein from Vibrio parahaemolyticus.
Vorobiev, S., Neely, H., Seetharaman, J., Patel, P., Xiao, R., Ciccosanti, C., Wang, H., Everett, J.K., Nair, R., Acton, T.B., Rost, B., Montelione, G.T., Tong, L., Hunt, J.F.To be published.