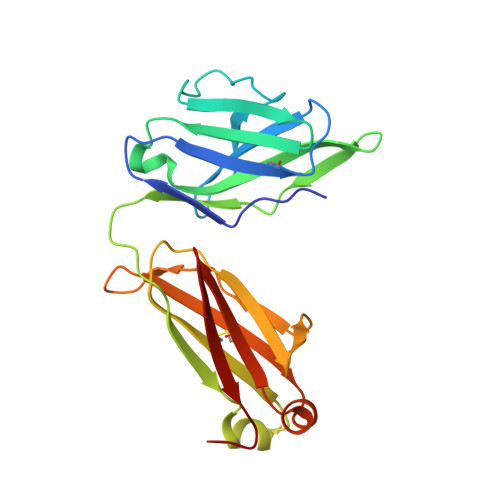
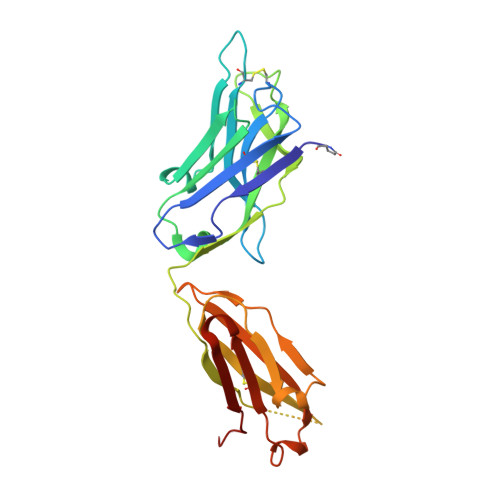
A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield.
Pejchal, R., Doores, K.J., Walker, L.M., Khayat, R., Huang, P.S., Wang, S.K., Stanfield, R.L., Julien, J.P., Ramos, A., Crispin, M., Depetris, R., Katpally, U., Marozsan, A., Cupo, A., Maloveste, S., Liu, Y., McBride, R., Ito, Y., Sanders, R.W., Ogohara, C., Paulson, J.C., Feizi, T., Scanlan, C.N., Wong, C.H., Moore, J.P., Olson, W.C., Ward, A.B., Poignard, P., Schief, W.R., Burton, D.R., Wilson, I.A.(2011) Science 334: 1097-1103
- PubMed: 21998254
- DOI: https://doi.org/10.1126/science.1213256
- Primary Citation of Related Structures:
3TV3, 3TWC, 3TYG - PubMed Abstract:
The HIV envelope (Env) protein gp120 is protected from antibody recognition by a dense glycan shield. However, several of the recently identified PGT broadly neutralizing antibodies appear to interact directly with the HIV glycan coat. Crystal structures of antigen-binding fragments (Fabs) PGT 127 and 128 with Man(9) at 1.65 and 1.29 angstrom resolution, respectively, and glycan binding data delineate a specific high mannose-binding site. Fab PGT 128 complexed with a fully glycosylated gp120 outer domain at 3.25 angstroms reveals that the antibody penetrates the glycan shield and recognizes two conserved glycans as well as a short β-strand segment of the gp120 V3 loop, accounting for its high binding affinity and broad specificity. Furthermore, our data suggest that the high neutralization potency of PGT 127 and 128 immunoglobulin Gs may be mediated by cross-linking Env trimers on the viral surface.
- Department of Molecular Biology, Skaggs Institute for Chemical Biology and International AIDS Vaccine Initiative (IAVI) Neutralizing Antibody Center, nhe Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: