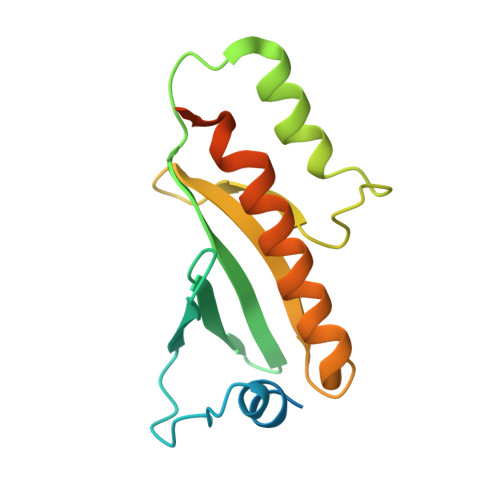
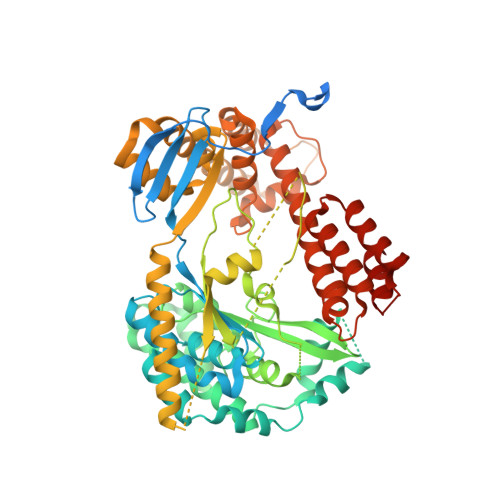
Structure of the Type III Secretion Effector Protein ExoU in Complex with Its Chaperone SpcU.
Halavaty, A.S., Borek, D., Tyson, G.H., Veesenmeyer, J.L., Shuvalova, L., Minasov, G., Otwinowski, Z., Hauser, A.R., Anderson, W.F.(2012) PLoS One 7: e49388-e49388
- PubMed: 23166655
- DOI: https://doi.org/10.1371/journal.pone.0049388
- Primary Citation of Related Structures:
3TU3 - PubMed Abstract:
Disease causing bacteria often manipulate host cells in a way that facilitates the infectious process. Many pathogenic gram-negative bacteria accomplish this by using type III secretion systems. In these complex secretion pathways, bacterial chaperones direct effector proteins to a needle-like secretion apparatus, which then delivers the effector protein into the host cell cytosol. The effector protein ExoU and its chaperone SpcU are components of the Pseudomonas aeruginosa type III secretion system. Secretion of ExoU has been associated with more severe infections in both humans and animal models. Here we describe the 1.92 Å X-ray structure of the ExoU-SpcU complex, a full-length type III effector in complex with its full-length cognate chaperone. Our crystallographic data allow a better understanding of the mechanism by which ExoU kills host cells and provides a foundation for future studies aimed at designing inhibitors of this potent toxin.
- Department of Molecular Pharmacology and Biological Chemistry, Feinberg School of Medicine, Northwestern University, Chicago, Illinois, USA.
Organizational Affiliation: